Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome
- PMID: 20207970
- PMCID: PMC2904004
- DOI: 10.1167/iovs.09-4853
Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome
Abstract
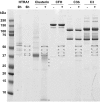
PURPOSE. To define the role of the serine protease HTRA1 in age-related macular degeneration (AMD) by examining its expression level and identifying its potential substrates in the context of primary RPE cell extracellular milieu. METHODS. Primary RPE cell cultures were established from human donor eyes and screened for CFH, ARMS2, and HTRA1 risk genotypes by using an allele-discrimination assay. HTRA1 expression in genotyped RPE cells was determined by using real-time PCR and quantitative proteomics. Potential HTRA1 substrates were identified by incubating RPE-conditioned medium with or without human recombinant HTRA1. Selectively cleaved proteins were quantified by using the differential stable isotope labeling by amino acids in cell culture (SILAC) strategy. RESULTS. HTRA1 mRNA levels were threefold higher in primary RPE cells homozygous for the HTRA1 promoter risk allele than in RPE cells with the wild-type allele, which translated into a twofold increase in HTRA1 secretion by RPE cells with the risk genotype. A total of 196 extracellular proteins were identified in the RPE secretome, and only 8 were found to be selectively cleaved by the human recombinant HTRA1. These include fibromodulin with 90% cleavage, clusterin (50%), ADAM9 (54%), vitronectin (54%), and alpha2-macroglobulin (55%), as well as some cell surface proteins including talin-1 (21%), fascin (40%), and chloride intracellular channel protein 1 (51%). CONCLUSIONS. Recombinant HTRA1 cleaves RPE-secreted proteins involved in regulation of the complement pathway (clusterin, vitronectin, and fibromodulin) and of amyloid deposition (clusterin, alpha2-macroglobulin, and ADAM9). These findings suggest a link between HTRA1, complement regulation, and amyloid deposition in AMD pathogenesis.
Figures





References
-
- Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science 2005;308:419–421 - PubMed
-
- Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science 2005;308:421–424 - PubMed
-
- Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet 2005;14:3227–3236 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

