Analysis of Lactobacillus sakei mutants selected after adaptation to the gastrointestinal tracts of axenic mice
- PMID: 20208026
- PMCID: PMC2863443
- DOI: 10.1128/AEM.02451-09
Analysis of Lactobacillus sakei mutants selected after adaptation to the gastrointestinal tracts of axenic mice
Abstract
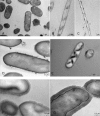
We recently showed that Lactobacillus sakei, a natural meat-borne lactic acid bacterium, can colonize the gastrointestinal tracts (GIT) of axenic mice but that this colonization in the intestinal environment selects L. sakei mutants showing modified colony morphology (small and rough) and cell shape, most probably resulting from the accumulation of various mutations that confer a selective advantage for persistence in the GIT. In the present study, we analyzed such clones, issued from three different L. sakei strains, in order to determine which functions were modified in the mutants. In the elongated filamentous cells of the rough clones, transmission electron microscopy (TEM) analysis showed a septation defect and dotted and slanted black bands, suggesting the presence of a helical structure around the cells. Comparison of the cytoplasmic and cell wall/membrane proteomes of the meat isolate L. sakei 23K and of one of its rough derivatives revealed a modified expression for 38 spots. The expression of six oxidoreductases, several stress proteins, and four ABC transporters was strongly reduced in the GIT-adapted strain, while the actin-like MreB protein responsible for cell shaping was upregulated. In addition, the expression of several enzymes involved in carbohydrate metabolism was modified, which may correlate with the observation of modified growth of mutants on various carbon sources. These results suggest that the modifications leading to a better adaptation to the GIT are pleiotropic and are characterized in a rough mutant by a different stress status, a cell wall modification, and modified use of energy sources, leading to an improved fitness for the colonization of the GIT.
Figures
Similar articles
-
Behavior of the meat-borne bacterium Lactobacillus sakei during its transit through the gastrointestinal tracts of axenic and conventional mice.Appl Environ Microbiol. 2009 Jul;75(13):4498-505. doi: 10.1128/AEM.02868-08. Epub 2009 May 15. Appl Environ Microbiol. 2009. PMID: 19447958 Free PMC article.
-
Catabolism of N-acetylneuraminic acid, a fitness function of the food-borne lactic acid bacterium Lactobacillus sakei, involves two newly characterized proteins.Appl Environ Microbiol. 2013 Mar;79(6):2012-8. doi: 10.1128/AEM.03301-12. Epub 2013 Jan 18. Appl Environ Microbiol. 2013. PMID: 23335758 Free PMC article.
-
Adaptive response of Lactobacillus sakei 23K during growth in the presence of meat extracts: a proteomic approach.Int J Food Microbiol. 2010 Aug 15;142(1-2):36-43. doi: 10.1016/j.ijfoodmicro.2010.05.014. Epub 2010 May 24. Int J Food Microbiol. 2010. PMID: 20580114
-
Adhesion properties of cell surface proteins in Lactobacillus strains in the GIT environment.Food Funct. 2022 Mar 21;13(6):3098-3109. doi: 10.1039/d1fo04328e. Food Funct. 2022. PMID: 35226005 Review.
-
Catabolic flexibility of mammalian-associated lactobacilli.Microb Cell Fact. 2013 May 16;12:48. doi: 10.1186/1475-2859-12-48. Microb Cell Fact. 2013. PMID: 23680304 Free PMC article. Review.
Cited by
-
Comparative genomics of Lactobacillus sakei with emphasis on strains from meat.Mol Genet Genomics. 2011 Apr;285(4):297-311. doi: 10.1007/s00438-011-0608-1. Epub 2011 Mar 3. Mol Genet Genomics. 2011. PMID: 21369871
-
Lactobacillus sakei: A Starter for Sausage Fermentation, a Protective Culture for Meat Products.Microorganisms. 2017 Sep 6;5(3):56. doi: 10.3390/microorganisms5030056. Microorganisms. 2017. PMID: 28878171 Free PMC article. Review.
-
Integrated phenotypic-genotypic approach to understand the influence of ultrasound on metabolic response of Lactobacillus sakei.PLoS One. 2018 Jan 25;13(1):e0191053. doi: 10.1371/journal.pone.0191053. eCollection 2018. PLoS One. 2018. PMID: 29370210 Free PMC article.
-
Correlation of Lactobacillus rhamnosus Genotypes and Carbohydrate Utilization Signatures Determined by Phenotype Profiling.Appl Environ Microbiol. 2015 Aug 15;81(16):5458-70. doi: 10.1128/AEM.00851-15. Epub 2015 Jun 5. Appl Environ Microbiol. 2015. PMID: 26048937 Free PMC article.
-
Global transcriptome response in Lactobacillus sakei during growth on ribose.BMC Microbiol. 2011 Jun 24;11:145. doi: 10.1186/1471-2180-11-145. BMC Microbiol. 2011. PMID: 21702908 Free PMC article.
References
-
- Alpert, C., J. Scheel, W. Engst, G. Loh, and M. Blaut. 2009. Adaptation of protein expression by Escherichia coli in the gastrointestinal tract of gnotobiotic mice. Environ. Microbiol. 11:751-761. - PubMed
-
- Berthier, F., M. Zagorec, M. Champomier-Vergès, S. D. Ehrlich, and F. Morel-Deville. 1996. Efficient transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. - PubMed
-
- Bredholt, S., and Nesbakken, T. 1999. Protective cultures inhibit growth of Listeria monocytogenes and Escherichia coli O157.H7 in cooked, vacuum and gas-packaged meat. Int. J. Food Microbiol. 53:43-52. - PubMed
-
- Chaillou, S., M. C. Champomier-Vergès, M. Cornet, A. M. Crutz-Le Coq, A. M. Dudez, V. Martin, S. Beaufils, E. Darbon-Rongère, R. Bossy, V. Loux, and M. Zagorec. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

