Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
- PMID: 20208152
- PMCID: PMC2833028
- DOI: 10.1107/S1744309109055158
Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Abstract
The trace-element oxyanion molybdate, which is required for the growth of many bacterial and archaeal species, is transported into the cell by an ATP-binding cassette (ABC) transporter superfamily uptake system called ModABC. ModABC consists of the ModA periplasmic solute-binding protein, the integral membrane-transport protein ModB and the ATP-binding and hydrolysis cassette protein ModC. In this study, X-ray crystal structures of ModA from the archaeon Methanosarcina acetivorans (MaModA) have been determined in the apoprotein conformation at 1.95 and 1.69 A resolution and in the molybdate-bound conformation at 2.25 and 2.45 A resolution. The overall domain structure of MaModA is similar to other ModA proteins in that it has a bilobal structure in which two mixed alpha/beta domains are linked by a hinge region. The apo MaModA is the first unliganded archaeal ModA structure to be determined: it exhibits a deep cleft between the two domains and confirms that upon binding ligand one domain is rotated towards the other by a hinge-bending motion, which is consistent with the 'Venus flytrap' model seen for bacterial-type periplasmic binding proteins. In contrast to the bacterial ModA structures, which have tetrahedral coordination of their metal substrates, molybdate-bound MaModA employs octahedral coordination of its substrate like other archaeal ModA proteins.
Figures
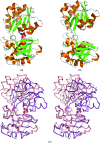

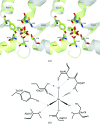

References
-
- Austermuhle, M. I., Hall, J. A., Klug, C. S. & Davidson, A. L. (2004). J. Biol. Chem.279, 28243–28250. - PubMed
-
- Balan, A., Santacruz-Perez, C., Moutran, A., Ferreira, L. C., Neshich, G. & Barbosa, J. A. (2008). Biochim. Biophys. Acta, 1784, 393–399. - PubMed
-
- Brylinski, M. & Skolnick, J. (2008). Proteins, 70, 363–377. - PubMed
-
- Clarke, T. E., Ku, S. Y., Dougan, D. R., Vogel, H. J. & Tari, L. W. (2000). Nature Struct. Biol.7, 287–291. - PubMed
-
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

