Activation of EGFR by proteasome inhibition requires HB-EGF in pancreatic cancer cells
- PMID: 20208558
- PMCID: PMC3134242
- DOI: 10.1038/onc.2010.52
Activation of EGFR by proteasome inhibition requires HB-EGF in pancreatic cancer cells
Abstract
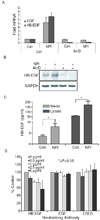
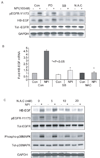
Resistance to drug treatments underlies the high lethality of pancreatic ductal adenocarcinoma. Along with others, we have recently identified that proteasome inhibition is a promising therapeutic option in this highly refractory disease. The pleiotropic effects of proteasome inhibition include the activation of apoptotic signaling pathways and also antiapoptotic signaling pathways such as EGFR, AKT and the MAP kinases that reduce the apoptotic potential of this class of drug. In this study, we sought to determine the mechanism behind the activation of EGFR in response to proteasome inhibition in pancreatic cancer cells. We found that the second-generation proteasome inhibitor NPI-0052 induced the mRNA transcription of several EGFR family ligands (EGF, HB-EGF and epiregulin), however only increases in HB-EGF were detected at the protein level. Using both pharmacological inhibitors and lentiviral-mediated shRNA knockdown of EGFR ligand expression, we discovered that ligand cleavage by MMP/ADAMs and HB-EGF expression is required for activation of EGFR in response to proteasome inhibition. Furthermore, we discover that induction of HB-EGF is dependent on reactive oxygen species and p38-MAPK signaling but not ERK and that the transcription factor SP-1 is involved in NPI-0052-induced HB-EGF transcription. Together, these results indicate that stress signaling leading to induction of HB-EGF expression and increases in MMP/ADAM-dependent HB-EGF cleavage are responsible for proteasome inhibitor-induced activation of EGFR in pancreatic cancer cells.
Figures




Similar articles
-
ERK1/2 mediate wounding- and G-protein-coupled receptor ligands-induced EGFR activation via regulating ADAM17 and HB-EGF shedding.Invest Ophthalmol Vis Sci. 2009 Jan;50(1):132-9. doi: 10.1167/iovs.08-2246. Epub 2008 Jul 24. Invest Ophthalmol Vis Sci. 2009. PMID: 18658095 Free PMC article.
-
Amphiregulin regulates the activation of ERK and Akt through epidermal growth factor receptor and HER3 signals involved in the progression of pancreatic cancer.Cancer Sci. 2010 Nov;101(11):2351-60. doi: 10.1111/j.1349-7006.2010.01671.x. Cancer Sci. 2010. PMID: 20726858 Free PMC article.
-
Metabolic regulation of EGFR effector and feedback signaling in pancreatic cancer cells requires K-Ras.Biochem Biophys Res Commun. 2020 Dec 10;533(3):424-428. doi: 10.1016/j.bbrc.2020.09.029. Epub 2020 Sep 21. Biochem Biophys Res Commun. 2020. PMID: 32972751 Free PMC article.
-
Fulvestrant regulates epidermal growth factor (EGF) family ligands to activate EGF receptor (EGFR) signaling in breast cancer cells.Breast Cancer Res Treat. 2013 Jun;139(2):351-60. doi: 10.1007/s10549-013-2541-y. Epub 2013 May 18. Breast Cancer Res Treat. 2013. PMID: 23686416 Free PMC article.
-
EGFR is dispensable for c-Met-mediated proliferation and survival activities in mouse adult liver oval cells.Cell Signal. 2012 Feb;24(2):505-513. doi: 10.1016/j.cellsig.2011.09.031. Epub 2011 Oct 7. Cell Signal. 2012. PMID: 22001397
Cited by
-
Phase 1 Study of Monotherapy with KHK2866, an Anti-Heparin-Binding Epidermal Growth Factor-Like Growth Factor Monoclonal Antibody, in Patients with Advanced Cancer.Target Oncol. 2016 Jun;11(3):317-27. doi: 10.1007/s11523-015-0394-5. Target Oncol. 2016. PMID: 26507836 Clinical Trial.
-
Autocrine expression of the epidermal growth factor receptor ligand heparin-binding EGF-like growth factor in cervical cancer.Int J Oncol. 2017 Jun;50(6):1947-1954. doi: 10.3892/ijo.2017.3980. Epub 2017 May 3. Int J Oncol. 2017. PMID: 28498437 Free PMC article.
-
High epiregulin expression in human U87 glioma cells relies on IRE1α and promotes autocrine growth through EGF receptor.BMC Cancer. 2013 Dec 13;13:597. doi: 10.1186/1471-2407-13-597. BMC Cancer. 2013. PMID: 24330607 Free PMC article.
-
s-HBEGF/SIRT1 circuit-dictated crosstalk between vascular endothelial cells and keratinocytes mediates sorafenib-induced hand-foot skin reaction that can be reversed by nicotinamide.Cell Res. 2020 Sep;30(9):779-793. doi: 10.1038/s41422-020-0309-6. Epub 2020 Apr 15. Cell Res. 2020. PMID: 32296111 Free PMC article.
-
Marizomib, a proteasome inhibitor for all seasons: preclinical profile and a framework for clinical trials.Curr Cancer Drug Targets. 2011 Mar;11(3):254-84. doi: 10.2174/156800911794519716. Curr Cancer Drug Targets. 2011. PMID: 21247382 Free PMC article. Review.
References
-
- Buck E, Eyzaguirre A, Haley JD, Gibson NW, Cagnoni P, Iwata KK. Inactivation of Akt by the epidermal growth factor receptor inhibitor erlotinib is mediated by HER-3 in pancreatic and colorectal tumor cell lines and contributes to erlotinib sensitivity. Mol Cancer Ther. 2006;5:2051–2059. - PubMed
-
- Codony-Servat J, Tapia MA, Bosch M, Oliva C, Domingo-Domenech J, Mellado B, et al. Differential cellular and molecular effects of bortezomib, a proteasome inhibitor, in human breast cancer cells. Mol Cancer Ther. 2006;5:665–675. - PubMed
-
- D'Addario M, Arora PD, McCulloch CA. Role of p38 in stress activation of Sp1. Gene. 2006;379:51–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

