The two stem cell microRNA gene clusters C19MC and miR-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas
- PMID: 20209130
- PMCID: PMC2831057
- DOI: 10.1371/journal.pone.0009485
The two stem cell microRNA gene clusters C19MC and miR-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas
Abstract
Thyroid adenomas are common benign human tumors with a high prevalence of about 5% of the adult population even in iodine sufficient areas. Rearrangements of chromosomal band 19q13.4 represent a frequent clonal cytogenetic deviation in these tumors making them the most frequent non-random chromosomal translocations in human epithelial tumors at all. Two microRNA (miRNA) gene clusters i.e. C19MC and miR-371-3 are located in close proximity to the breakpoint region of these chromosomal rearrangements and have been checked for a possible up-regulation due to the genomic alteration. In 4/5 cell lines established from thyroid adenomas with 19q13.4 rearrangements and 5/5 primary adenomas with that type of rearrangement both the C19MC and miR-371-3 cluster were found to be significantly overexpressed compared to controls lacking that particular chromosome abnormality. In the remaining cell line qRT-PCR revealed overexpression of members of the miR-371-3 cluster only which might be due to a deletion accompanying the chromosomal rearrangement in that case. In depth molecular characterization of the breakpoint in a cell line from one adenoma of this type reveals the existence of large Pol-II mRNA fragments as the most likely source of up-regulation of the C19MC cluster. The up-regulation of the clusters is likely to be causally associated with the pathogenesis of the corresponding tumors. Of note, the expression of miRNAs miR-520c and miR-373 is known to characterize stem cells and in terms of molecular oncology has been implicated in invasive growth of epithelial cells in vitro and in vivo thus allowing to delineate a distinct molecular subtype of thyroid adenomas. Besides thyroid adenomas rearrangements of 19q13.4 are frequently found in other human neoplasias as well, suggesting that activation of both clusters might be a more general phenomenon in human neoplasias.
Conflict of interest statement
Figures






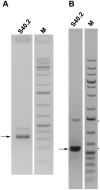

References
-
- DeLellis RA. Lyon: IARC Press; 2004. Pathology and genetics of tumours of endocrine organs: [… editorial and consensus conference in Lyon, France, April 23 - 26, 2003]. 320 S. p.
-
- Belge G, Roque L, Soares J, Bruckmann S, Thode B, et al. Cytogenetic investigations of 340 thyroid hyperplasias and adenomas revealing correlations between cytogenetic findings and histology. Cancer Genet Cytogenet. 1998;101:42–48. - PubMed
-
- Belge G, Rippe V, Meiboom M, Drieschner N, Garcia E, et al. Delineation of a 150-kb breakpoint cluster in benign thyroid tumors with 19q13.4 aberrations. Cytogenet Cell Genet. 2001;93:48–51. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

