Complete in vitro life cycle of Trypanosoma congolense: development of genetic tools
- PMID: 20209144
- PMCID: PMC2830455
- DOI: 10.1371/journal.pntd.0000618
Complete in vitro life cycle of Trypanosoma congolense: development of genetic tools
Abstract
Background: Animal African trypanosomosis, a disease mainly caused by the protozoan parasite Trypanosoma congolense, is a major constraint to livestock productivity and has a significant impact in the developing countries of Africa. RNA interference (RNAi) has been used to study gene function and identify drug and vaccine targets in a variety of organisms including trypanosomes. However, trypanosome RNAi studies have mainly been conducted in T. brucei, as a model for human infection, largely ignoring livestock parasites of economical importance such as T. congolense, which displays different pathogenesis profiles. The whole T. congolense life cycle can be completed in vitro, but this attractive model displayed important limitations: (i) genetic tools were currently limited to insect forms and production of modified infectious BSF through differentiation was never achieved, (ii) in vitro differentiation techniques lasted several months, (iii) absence of long-term bloodstream forms (BSF) in vitro culture prevented genomic analyses.
Methodology/principal findings: We optimized culture conditions for each developmental stage and secured the differentiation steps. Specifically, we devised a medium adapted for the strenuous development of stable long-term BSF culture. Using Amaxa nucleofection technology, we greatly improved the transfection rate of the insect form and designed an inducible transgene expression system using the IL3000 reference strain. We tested it by expression of reporter genes and through RNAi. Subsequently, we achieved the complete in vitro life cycle with dramatically shortened time requirements for various wild type and transgenic strains. Finally, we established the use of modified strains for experimental infections and underlined a host adaptation phase requirement.
Conclusions/significance: We devised an improved T. congolense model, which offers the opportunity to perform functional genomics analyses throughout the whole life cycle. It represents a very useful tool to understand pathogenesis mechanisms and to study potential therapeutic targets either in vitro or in vivo using a mouse model.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



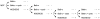

 ), or in absence of reducing agents (Bathocuproin, 2-mercaptoethanol and cystein were removed) (——▪). Cells were counted every 24 h.
), or in absence of reducing agents (Bathocuproin, 2-mercaptoethanol and cystein were removed) (——▪). Cells were counted every 24 h.
References
-
- Kappmeier K, Nevill EM, Bagnall RJ. Review of tsetse flies and trypanosomosis in South Africa. Onderstepoort J Vet Res. 1998;65:195–203. - PubMed
-
- Van den Bossche P. Some general aspects of the distribution and epidemiology of bovine trypanosomosis in southern Africa. Int J Parasitol. 2001;31:592–598. - PubMed
-
- LaCount DJ, Donelson JE. RNA interference in African trypanosomes. Protist. 2001;152:103–111. - PubMed
-
- Wirtz E, Clayton C. Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science. 1995;268:1179–1183. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

