Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity
- PMID: 20211136
- PMCID: PMC2875855
- DOI: 10.1016/j.cell.2010.01.038
Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity
Abstract
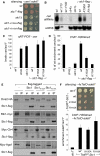
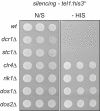
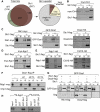
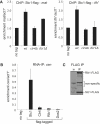
In fission yeast, RNAi directs heterochromatin formation at centromeres, telomeres, and the mating type locus. Noncoding RNAs transcribed from repeat elements generate siRNAs that are incorporated into the Argonaute-containing RITS complex and direct it to nascent homologous transcripts. This leads to recruitment of the CLRC complex, including the histone methyltransferase Clr4, promoting H3K9 methylation and heterochromatin formation. A key question is what mediates the recruitment of Clr4/CLRC to transcript-bound RITS. We have identified a LIM domain protein, Stc1, that is required for centromeric heterochromatin integrity. Our analyses show that Stc1 is specifically required to establish H3K9 methylation via RNAi, and interacts both with the RNAi effector Ago1, and with the chromatin-modifying CLRC complex. Moreover, tethering Stc1 to a euchromatic locus is sufficient to induce silencing and heterochromatin formation independently of RNAi. We conclude that Stc1 associates with RITS on centromeric transcripts and recruits CLRC, thereby coupling RNAi to chromatin modification.
(c) 2010 Elsevier Inc. All rights reserved.
Figures

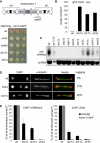
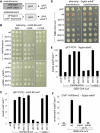

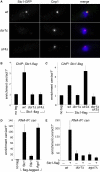
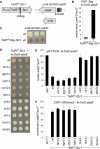



References
-
- Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. - PubMed
-
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., 3rd, Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

