Cloning whole bacterial genomes in yeast
- PMID: 20211840
- PMCID: PMC2860123
- DOI: 10.1093/nar/gkq119
Cloning whole bacterial genomes in yeast
Abstract
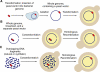
Most microbes have not been cultured, and many of those that are cultivatable are difficult, dangerous or expensive to propagate or are genetically intractable. Routine cloning of large genome fractions or whole genomes from these organisms would significantly enhance their discovery and genetic and functional characterization. Here we report the cloning of whole bacterial genomes in the yeast Saccharomyces cerevisiae as single-DNA molecules. We cloned the genomes of Mycoplasma genitalium (0.6 Mb), M. pneumoniae (0.8 Mb) and M. mycoides subspecies capri (1.1 Mb) as yeast circular centromeric plasmids. These genomes appear to be stably maintained in a host that has efficient, well-established methods for DNA manipulation.
Figures
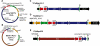

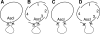
References
-
- Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods. 2008;5:41–43. - PubMed
-
- Yonemura I, Nakada K, Sato A, Hayashi J, Fujita K, Kaneko S, Itaya M. Direct cloning of full-length mouse mitochondrial DNA using a Bacillus subtilis genome vector. Gene. 2007;391:171–177. - PubMed
-
- Wheeler VC, Aitken M, Coutelle C. Modification of the mouse mitochondrial genome by insertion of an exogenous gene. Gene. 1997;198:203–209. - PubMed
-
- Gupta M, Hoo B. Entire maize chloroplast genome is stably maintained in a yeast artificial chromosome. Plant Mol. Biol. 1991;17:361–369. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

