FXR regulates liver repair after CCl4-induced toxic injury
- PMID: 20211986
- PMCID: PMC3284774
- DOI: 10.1210/me.2009-0286
FXR regulates liver repair after CCl4-induced toxic injury
Abstract
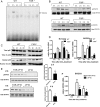
Liver repair is key to resuming homeostasis and preventing fibrogenesis as well as other liver diseases. Farnesoid X receptor (FXR, NR1H4) is an emerging liver metabolic regulator and cell protector. Here we show that FXR is essential to promote liver repair after carbon tetrachloride (CCl(4))-induced injury. Expression of hepatic FXR in wild-type mice was strongly suppressed by CCl(4) treatment, and bile acid homeostasis was disrupted. Liver injury was induced in both wild-type and FXR(-/-) mice by CCl(4), but FXR(-/-) mice had more severe defects in liver repair than wild-type mice. FXR(-/-) livers had a decreased peak of regenerative DNA synthesis and reduced induction of genes involved in liver regeneration. Moreover, FXR(-/-) mice displayed increased mortality and enhanced hepatocyte deaths. During the early stages of liver repair after CCl(4) treatment, we observed overproduction of TNFalpha and a strong decrease of phosphorylation and DNA-binding activity of signal transducer and activator of transcription 3 in livers from FXR(-/-) mice. Exogenous expression of a constitutively active signal transducer and activator of transcription 3 protein in FXR(-/-) liver effectively reduced hepatocyte death and liver injury after CCl(4) treatment. These results suggest that FXR is required to regulate normal liver repair by promoting regeneration and preventing cell death.
Figures







References
-
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C 1995 Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687–693 - PubMed
-
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B 1999 Identification of a nuclear receptor for bile acids. Science 284:1362–1365 - PubMed
-
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM 1999 Bile acids: natural ligands for an orphan nuclear receptor. Science 284:1365–1368 - PubMed
-
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM 1999 Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell 3:543–553 - PubMed
-
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA 2000 A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 6:517–526 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

