GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome
- PMID: 20212525
- PMCID: PMC2858444
- DOI: 10.1038/msb.2010.8
GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome
Abstract
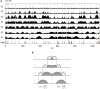
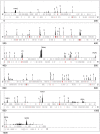
Previous studies have led to a picture wherein the replication of DNA progresses at variable rates over different parts of the budding yeast genome. These prior experiments, focused on production of nascent DNA, have been interpreted to imply that the dynamics of replication fork progression are strongly affected by local chromatin structure/architecture, and by interaction with machineries controlling transcription, repair and epigenetic maintenance. Here, we adopted a complementary approach for assaying replication dynamics using whole genome time-resolved chromatin immunoprecipitation combined with microarray analysis of the GINS complex, an integral member of the replication fork. Surprisingly, our data show that this complex progresses at highly uniform rates regardless of genomic location, revealing that replication fork dynamics in yeast is simpler and more uniform than previously envisaged. In addition, we show how the synergistic use of experiment and modeling leads to novel biological insights. In particular, a parsimonious model allowed us to accurately simulate fork movement throughout the genome and also revealed a subtle phenomenon, which we interpret as arising from low-frequency fork arrest.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes.Genes Dev. 2006 Nov 15;20(22):3104-16. doi: 10.1101/gad.1478906. Genes Dev. 2006. PMID: 17114583 Free PMC article.
-
Replication dynamics of the yeast genome.Science. 2001 Oct 5;294(5540):115-21. doi: 10.1126/science.294.5540.115. Science. 2001. PMID: 11588253
-
A variable fork rate affects timing of origin firing and S phase dynamics in Saccharomyces cerevisiae.J Biotechnol. 2013 Oct 20;168(2):174-84. doi: 10.1016/j.jbiotec.2013.06.022. Epub 2013 Jul 9. J Biotechnol. 2013. PMID: 23850861
-
Genome-wide patterns of histone modifications in yeast.Nat Rev Mol Cell Biol. 2006 Sep;7(9):657-66. doi: 10.1038/nrm1986. Epub 2006 Aug 16. Nat Rev Mol Cell Biol. 2006. PMID: 16912715 Review.
-
The dynamics of chromosome replication in yeast.Curr Top Dev Biol. 2003;55:1-73. doi: 10.1016/s0070-2153(03)01001-9. Curr Top Dev Biol. 2003. PMID: 12959193 Review. No abstract available.
Cited by
-
Physical tethering and volume exclusion determine higher-order genome organization in budding yeast.Genome Res. 2012 Jul;22(7):1295-305. doi: 10.1101/gr.129437.111. Epub 2012 May 22. Genome Res. 2012. PMID: 22619363 Free PMC article.
-
The torsional state of DNA within the chromosome.Chromosoma. 2011 Aug;120(4):323-34. doi: 10.1007/s00412-011-0324-y. Epub 2011 May 13. Chromosoma. 2011. PMID: 21567156 Review.
-
Model-based analysis of DNA replication profiles: predicting replication fork velocity and initiation rate by profiling free-cycling cells.Genome Res. 2017 Feb;27(2):310-319. doi: 10.1101/gr.205849.116. Epub 2016 Dec 27. Genome Res. 2017. PMID: 28028072 Free PMC article.
-
The multiple activations in budding yeast S-phase checkpoint are Poisson processes.PNAS Nexus. 2023 Oct 26;2(11):pgad342. doi: 10.1093/pnasnexus/pgad342. eCollection 2023 Nov. PNAS Nexus. 2023. PMID: 37941810 Free PMC article.
-
Do replication forks control late origin firing in Saccharomyces cerevisiae?Nucleic Acids Res. 2012 Mar;40(5):2010-9. doi: 10.1093/nar/gkr982. Epub 2011 Nov 15. Nucleic Acids Res. 2012. PMID: 22086957 Free PMC article.
References
-
- Deshpande A, Newlon C (1996) DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

