Design and utilization of epitope-based databases and predictive tools
- PMID: 20213141
- PMCID: PMC2843836
- DOI: 10.1007/s00251-010-0435-2
Design and utilization of epitope-based databases and predictive tools
Abstract
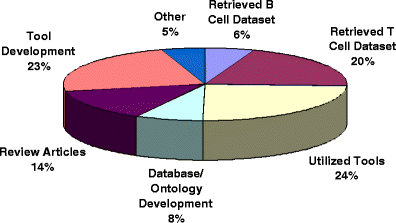
In the last decade, significant progress has been made in expanding the scope and depth of publicly available immunological databases and online analysis resources, which have become an integral part of the repertoire of tools available to the scientific community for basic and applied research. Herein, we present a general overview of different resources and databases currently available. Because of our association with the Immune Epitope Database and Analysis Resource, this resource is reviewed in more detail. Our review includes aspects such as the development of formal ontologies and the type and breadth of analytical tools available to predict epitopes and analyze immune epitope data. A common feature of immunological databases is the requirement to host large amounts of data extracted from disparate sources. Accordingly, we discuss and review processes to curate the immunological literature, as well as examples of how the curated data can be used to generate a meta-analysis of the epitope knowledge currently available for diseases of worldwide concern, such as influenza and malaria. Finally, we review the impact of immunological databases, by analyzing their usage and citations, and by categorizing the type of citations. Taken together, the results highlight the growing impact and utility of immunological databases for the scientific community.
Figures
Similar articles
-
Resource Disambiguator for the Web: Extracting Biomedical Resources and Their Citations from the Scientific Literature.PLoS One. 2016 Jan 5;11(1):e0146300. doi: 10.1371/journal.pone.0146300. eCollection 2016. PLoS One. 2016. PMID: 26730820 Free PMC article.
-
The immune epitope database: a historical retrospective of the first decade.Immunology. 2012 Oct;137(2):117-23. doi: 10.1111/j.1365-2567.2012.03611.x. Immunology. 2012. PMID: 22681406 Free PMC article.
-
Curation of complex, context-dependent immunological data.BMC Bioinformatics. 2006 Jul 12;7:341. doi: 10.1186/1471-2105-7-341. BMC Bioinformatics. 2006. PMID: 16836764 Free PMC article.
-
Immunoinformatics comes of age.PLoS Comput Biol. 2006 Jun 30;2(6):e71. doi: 10.1371/journal.pcbi.0020071. PLoS Comput Biol. 2006. PMID: 16846250 Free PMC article. Review.
-
The Immune Epitope Database and Analysis Resource Program 2003-2018: reflections and outlook.Immunogenetics. 2020 Feb;72(1-2):57-76. doi: 10.1007/s00251-019-01137-6. Epub 2019 Nov 25. Immunogenetics. 2020. PMID: 31761977 Free PMC article. Review.
Cited by
-
Bridging Computational Vaccinology and Vaccine Development Through Systematic Identification, Characterization, and Downselection of Conserved and Variable Circumsporozoite Protein CD4 T Cell Epitopes From Diverse Plasmodium falciparum Strains.Front Immunol. 2021 Jun 8;12:689920. doi: 10.3389/fimmu.2021.689920. eCollection 2021. Front Immunol. 2021. PMID: 34168657 Free PMC article.
-
Driving forces of proteasome-catalyzed peptide splicing in yeast and humans.Mol Cell Proteomics. 2012 Oct;11(10):1008-23. doi: 10.1074/mcp.M112.020164. Epub 2012 Jul 20. Mol Cell Proteomics. 2012. PMID: 22822185 Free PMC article.
-
Proteomics for Development of Food Allergy Vaccines.Methods Mol Biol. 2022;2410:673-689. doi: 10.1007/978-1-0716-1884-4_36. Methods Mol Biol. 2022. PMID: 34914075
-
Identification of critical amino acids in an immunodominant IgE epitope of Pen c 13, a major allergen from Penicillium citrinum.PLoS One. 2012;7(4):e34627. doi: 10.1371/journal.pone.0034627. Epub 2012 Apr 10. PLoS One. 2012. PMID: 22506037 Free PMC article.
-
Human CD4+ T cell response to human herpesvirus 6.J Virol. 2012 May;86(9):4776-92. doi: 10.1128/JVI.06573-11. Epub 2012 Feb 22. J Virol. 2012. PMID: 22357271 Free PMC article.
References
-
- Aurrecoechea C, Heiges M, Wang H, Wang Z, Fischer S, Rhodes P, Miller J, Kraemer E, Stoeckert CJ, Jr, Roos DS, Kissinger JC. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 2007;35(Database issue):D427–D430. doi: 10.1093/nar/gkl880. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases