Cholesterol-binding viral proteins in virus entry and morphogenesis
- PMID: 20213541
- PMCID: PMC7176229
- DOI: 10.1007/978-90-481-8622-8_3
Cholesterol-binding viral proteins in virus entry and morphogenesis
Abstract
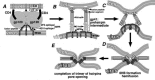
Up to now less than a handful of viral cholesterol-binding proteins have been characterized, in HIV, influenza virus and Semliki Forest virus. These are proteins with roles in virus entry or morphogenesis. In the case of the HIV fusion protein gp41 cholesterol binding is attributed to a cholesterol recognition consensus (CRAC) motif in a flexible domain of the ectodomain preceding the trans-membrane segment. This specific CRAC sequence mediates gp41 binding to a cholesterol affinity column. Mutations in this motif arrest virus fusion at the hemifusion stage and modify the ability of the isolated CRAC peptide to induce segregation of cholesterol in artificial membranes.Influenza A virus M2 protein co-purifies with cholesterol. Its proton translocation activity, responsible for virus uncoating, is not cholesterol-dependent, and the transmembrane channel appears too short for integral raft insertion. Cholesterol binding may be mediated by CRAC motifs in the flexible post-TM domain, which harbours three determinants of binding to membrane rafts. Mutation of the CRAC motif of the WSN strain attenuates virulence for mice. Its affinity to the raft-non-raft interface is predicted to target M2 protein to the periphery of lipid raft microdomains, the sites of virus assembly. Its influence on the morphology of budding virus implicates M2 as factor in virus fission at the raft boundary. Moreover, M2 is an essential factor in sorting the segmented genome into virus particles, indicating that M2 also has a role in priming the outgrowth of virus buds.SFV E1 protein is the first viral type-II fusion protein demonstrated to directly bind cholesterol when the fusion peptide loop locks into the target membrane. Cholesterol binding is modulated by another, proximal loop, which is also important during virus budding and as a host range determinant, as shown by mutational studies.
Figures







References
-
- Ali A., Nayak D.P. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology. 2000;276:289–303. - PubMed
-
- Asano K., Asano A. Binding of cholesterol and inhibitory peptide derivatives with the fusogenic hydrophobic sequence of F-glycoprotein of HVJ (Sendai virus): possible implication in the fusion reaction. Biochemistry. 1988;27:1321–1329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

