Relationship of common candidate gene variants to electrocardiographic T-wave peak to T-wave end interval and T-wave morphology parameters
- PMID: 20215044
- PMCID: PMC2904845
- DOI: 10.1016/j.hrthm.2010.03.002
Relationship of common candidate gene variants to electrocardiographic T-wave peak to T-wave end interval and T-wave morphology parameters
Abstract
Background: Single-nucleotide polymorphisms (SNPs) in genes encoding cardiac ion channels and nitric oxide synthase-1 adaptor protein (NOS1AP) are associated with electrocardiographic (ECG) QT-interval duration, but the association of these SNPs with new, prognostically important ECG measures of ventricular repolarization is unknown.
Objective: The purpose of this study was to examine the relationship of SNPs to ECG T-wave peak to T-wave end (TPE) interval and T-wave morphology parameters.
Methods: We studied 5,890 adults attending the Health 2000 Study, a Finnish epidemiologic survey. TPE interval and four T-wave morphology parameters were measured from digital 12-lead ECGs and related to the seven SNPs showing a phenotypic effect on QT-interval duration in the Health 2000 Study population.
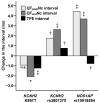
Results: In multivariable analyses, the KCNH2 K897T minor allele was associated with a 1.2-ms TPE-interval shortening (P = .00005) and the KCNH2 intronic rs3807375 minor allele was associated with a 0.8-ms TPE-interval prolongation (P = .001), whereas the KCNE1 D85N variant had no TPE-interval effect (P = .20). NOS1AP minor alleles (rs2880058, rs4657139, rs10918594, rs10494366) were associated with a shorter TPE interval (effects from 0.5 to 0.8 ms, P from .032 to .002), which resulted from their stronger effects on QT(peak) than QT(end) interval. None of the SNPs showed a consistent association with T-wave morphology parameters.
Conclusion: KCNH2 K897T and rs3807375 as well as the four studied NOS1AP variants have modest effects on ECG TPE interval but are not related to T-wave morphology measures. The previously observed prognostic value of T-wave morphology parameters likely is not based on these SNPs.
Copyright 2010 Heart Rhythm Society. Published by Elsevier Inc. All rights reserved.
Figures
Comment in
-
Genetic variants and ECG pattern variability in long QT syndrome: how far are we?Heart Rhythm. 2010 Jul;7(7):904-5. doi: 10.1016/j.hrthm.2010.04.015. Epub 2010 Apr 18. Heart Rhythm. 2010. PMID: 20406697 No abstract available.
Similar articles
-
Common candidate gene variants are associated with QT interval duration in the general population.J Intern Med. 2009 Apr;265(4):448-58. doi: 10.1111/j.1365-2796.2008.02026.x. Epub 2009 Oct 25. J Intern Med. 2009. PMID: 19019189 Free PMC article.
-
Common variation in NOS1AP and KCNH2 genes and QT interval duration in young adults. The Cardiovascular Risk in Young Finns Study.Ann Med. 2009;41(2):144-51. doi: 10.1080/07853890802392529. Ann Med. 2009. PMID: 18785031
-
Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study.Circulation. 2007 Jul 3;116(1):10-6. doi: 10.1161/CIRCULATIONAHA.106.676783. Epub 2007 Jun 18. Circulation. 2007. PMID: 17576865
-
Functional significance of KCNH2 (HERG) K897T polymorphism for cardiac repolarization assessed by analysis of T-wave morphology.Ann Noninvasive Electrocardiol. 2006 Jan;11(1):57-62. doi: 10.1111/j.1542-474X.2006.00083.x. Ann Noninvasive Electrocardiol. 2006. PMID: 16472284 Free PMC article.
-
Systematic Meta-Analysis of the Association Between a Common NOS1AP Genetic Polymorphism, the QTc Interval, and Sudden Death.Int Heart J. 2019 Sep 27;60(5):1083-1090. doi: 10.1536/ihj.19-024. Epub 2019 Aug 23. Int Heart J. 2019. PMID: 31447468
Cited by
-
Clinical and genetic determinants of torsade de pointes risk.Circulation. 2012 Apr 3;125(13):1684-94. doi: 10.1161/CIRCULATIONAHA.111.080887. Circulation. 2012. PMID: 22474311 Free PMC article. Review. No abstract available.
-
KCNE genetics and pharmacogenomics in cardiac arrhythmias: much ado about nothing?Expert Rev Clin Pharmacol. 2013 Jan;6(1):49-60. doi: 10.1586/ecp.12.76. Expert Rev Clin Pharmacol. 2013. PMID: 23272793 Free PMC article. Review.
-
The Relationship between Angiotensin-II Type 1 Receptor Gene Polymorphism and Repolarization Parameters after a First Anterior Acute Myocardial Infarction.Korean Circ J. 2016 Nov;46(6):791-797. doi: 10.4070/kcj.2016.46.6.791. Epub 2016 Nov 1. Korean Circ J. 2016. PMID: 27826337 Free PMC article.
-
Common Genetic Variants Modulate the Electrocardiographic Tpeak-to-Tend Interval.Am J Hum Genet. 2020 Jun 4;106(6):764-778. doi: 10.1016/j.ajhg.2020.04.009. Epub 2020 May 7. Am J Hum Genet. 2020. PMID: 32386560 Free PMC article.
-
Predisposition of functional genetic variants of A-kinase anchoring protein 10 toward acquired repolarization disorders in high-risk vascular surgery patients.Ther Clin Risk Manag. 2018 Jul 26;14:1315-1322. doi: 10.2147/TCRM.S167086. eCollection 2018. Ther Clin Risk Manag. 2018. PMID: 30100729 Free PMC article.
References
-
- Noseworthy PA, Newton-Cheh C. Genetic determinants of sudden cardiac death. Circulation. 2008;118:1854–1863. - PubMed
-
- Bezzina CR, Verkerk AO, Busjahn A, et al. A common polymorphism in KCNH2 (HERG) hastens cardiac repolarization. Cardiovasc Res. 2003;59:27–36. - PubMed
-
- Gouas L, Nicaud V, Berthet M, et al. Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. Eur J Hum Genet. 2005;13:1213–1222. - PubMed
-
- Pfeufer A, Jalilzadeh S, Perz S, et al. Common variants in myocardial ion channel genes modify the QT interval in the general population: results from the KORA study. Circ Res. 2005;96:693–701. - PubMed
-
- Newton-Cheh C, Guo CY, Larson MG, et al. Common genetic variation in KCNH2 is associated with QT interval duration: the Framingham Heart Study. Circulation. 2007;116:1128–1136. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

