Controlled expression of branch-forming mannosyltransferase is critical for mycobacterial lipoarabinomannan biosynthesis
- PMID: 20215111
- PMCID: PMC2859491
- DOI: 10.1074/jbc.M109.077297
Controlled expression of branch-forming mannosyltransferase is critical for mycobacterial lipoarabinomannan biosynthesis
Abstract
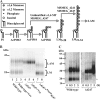
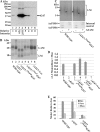
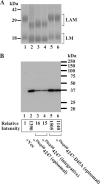
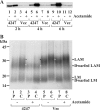
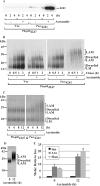
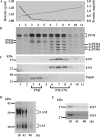
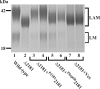
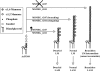
Lipomannan (LM) and lipoarabinomannan (LAM) are phosphatidylinositol-anchored glycans present in the mycobacterial cell wall. In Mycobacterium smegmatis, the mannan core of LM/LAM constitutes a linear chain of 20-25 alpha1,6-mannoses elaborated by 8-9 alpha1,2-monomannose side branches. At least two alpha1,6-mannosyltransferases mediate the linear mannose chain elongation, and one branching alpha1,2-mannosyltransferase (encoded by MSMEG_4247) transfers monomannose branches. An MSMEG_4247 deletion mutant accumulates branchless LAM and interestingly fails to accumulate LM, suggesting an unexpected role of mannose branching for LM synthesis or maintenance. To understand the roles of MSMEG_4247-mediated branching more clearly, we analyzed the MSMEG_4247 deletion mutant in detail. Our study showed that the deletion mutant restored the synthesis of wild-type LM and LAM upon the expression of MSMEG_4247 at wild-type levels. In striking contrast, overexpression of MSMEG_4247 resulted in the accumulation of dwarfed LM/LAM, although monomannose branching was restored. The dwarfed LAM carried a mannan chain less than half the length of wild-type LAM and was elaborated by an arabinan that was about 4 times smaller. Induced overexpression of an elongating alpha1,6-mannosyltransferase competed with the overexpressed branching enzyme, alleviating the dwarfing effect of the branching enzyme. In wild-type cells, LM and LAM decreased in quantity in the stationary phase, and the expression levels of branching and elongating mannosyltransferases were reduced in concert, presumably to avoid producing abnormal LM/LAM. These data suggest that the coordinated expressions of branching and elongating mannosyltransferases are critical for mannan backbone elongation.
Figures
Similar articles
-
Role of LmeA, a Mycobacterial Periplasmic Protein, in Maintaining the Mannosyltransferase MptA and Its Product Lipomannan under Stress.mSphere. 2020 Nov 4;5(6):e01039-20. doi: 10.1128/mSphere.01039-20. mSphere. 2020. PMID: 33148829 Free PMC article.
-
Biosynthesis of mycobacterial lipoarabinomannan: role of a branching mannosyltransferase.Proc Natl Acad Sci U S A. 2006 Sep 12;103(37):13664-9. doi: 10.1073/pnas.0603049103. Epub 2006 Aug 31. Proc Natl Acad Sci U S A. 2006. PMID: 16945913 Free PMC article.
-
A single arabinan chain is attached to the phosphatidylinositol mannosyl core of the major immunomodulatory mycobacterial cell envelope glycoconjugate, lipoarabinomannan.J Biol Chem. 2014 Oct 31;289(44):30249-30256. doi: 10.1074/jbc.M114.599415. Epub 2014 Sep 17. J Biol Chem. 2014. PMID: 25231986 Free PMC article.
-
Lipoarabinomannan of Mycobacterium: mannose capping by a multifunctional terminal mannosyltransferase.Proc Natl Acad Sci U S A. 2008 Nov 18;105(46):17973-7. doi: 10.1073/pnas.0807761105. Epub 2008 Nov 12. Proc Natl Acad Sci U S A. 2008. PMID: 19004785 Free PMC article.
-
Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response.Mol Microbiol. 2004 Jul;53(2):391-403. doi: 10.1111/j.1365-2958.2004.04183.x. Mol Microbiol. 2004. PMID: 15228522 Review.
Cited by
-
Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis.mBio. 2013 Feb 19;4(1):e00472-12. doi: 10.1128/mBio.00472-12. mBio. 2013. PMID: 23422411 Free PMC article.
-
An essential periplasmic protein coordinates lipid trafficking and is required for asymmetric polar growth in mycobacteria.Elife. 2022 Nov 8;11:e80395. doi: 10.7554/eLife.80395. Elife. 2022. PMID: 36346214 Free PMC article.
-
Fluorescence Imaging-Based Discovery of Membrane Domain-Associated Proteins in Mycobacterium smegmatis.J Bacteriol. 2021 Oct 25;203(22):e0041921. doi: 10.1128/JB.00419-21. Epub 2021 Sep 13. J Bacteriol. 2021. PMID: 34516286 Free PMC article.
-
Acid-Fast Positive and Acid-Fast Negative Mycobacterium tuberculosis: The Koch Paradox.Microbiol Spectr. 2017 Mar;5(2):10.1128/microbiolspec.tbtb2-0003-2015. doi: 10.1128/microbiolspec.TBTB2-0003-2015. Microbiol Spectr. 2017. PMID: 28337966 Free PMC article. Review.
-
Tuberculostearic Acid Controls Mycobacterial Membrane Compartmentalization.mBio. 2023 Apr 25;14(2):e0339622. doi: 10.1128/mbio.03396-22. Epub 2023 Mar 28. mBio. 2023. PMID: 36976029 Free PMC article.
References
-
- Brennan P. J. (2003) Tuberculosis 83, 91–97 - PubMed
-
- Briken V., Porcelli S. A., Besra G. S., Kremer L. (2004) Mol. Microbiol. 53, 391–403 - PubMed
-
- Alderwick L. J., Birch H. L., Mishra A. K., Eggeling L., Besra G. S. (2007) Biochem. Soc. Trans. 35, 1325–1328 - PubMed
-
- Khoo K. H., Dell A., Morris H. R., Brennan P. J., Chatterjee D. (1995) Glycobiology 5, 117–127 - PubMed
-
- Lee Y. C., Ballou C. E. (1964) J. Biol. Chem. 239, 1316–1327 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

