MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing
- PMID: 20215419
- PMCID: PMC2882868
- DOI: 10.1093/molehr/gaq017
MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing
Abstract
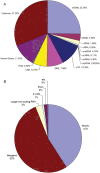
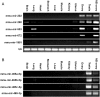
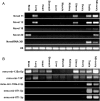
Small non-coding RNAs, such as microRNAs (miRNAs), are involved in diverse biological processes including organ development and tissue differentiation. Global disruption of miRNA biogenesis in Dicer knockout mice disrupts early embryogenesis and primordial germ cell formation. However, the role of miRNAs in early folliculogenesis is poorly understood. In order to identify a full transcriptome set of small RNAs expressed in the newborn (NB) ovary, we extracted small RNA fraction from mouse NB ovary tissues and subjected it to massive parallel sequencing using the Genome Analyzer from Illumina. Massive sequencing produced 4 655 992 reads of 33 bp each representing a total of 154 Mbp of sequence data. The Pash alignment algorithm mapped 50.13% of the reads to the mouse genome. Sequence reads were clustered based on overlapping mapping coordinates and intersected with known miRNAs, small nucleolar RNAs (snoRNAs), piwi-interacting RNA (piRNA) clusters and repetitive genomic regions; 25.2% of the reads mapped to known miRNAs, 25.5% to genomic repeats, 3.5% to piRNAs and 0.18% to snoRNAs. Three hundred and ninety-eight known miRNA species were among the sequenced small RNAs, and 118 isomiR sequences that are not in the miRBase database. Let-7 family was the most abundantly expressed miRNA, and mmu-mir-672, mmu-mir-322, mmu-mir-503 and mmu-mir-465 families are the most abundant X-linked miRNA detected. X-linked mmu-mir-503, mmu-mir-672 and mmu-mir-465 family showed preferential expression in testes and ovaries. We also identified four novel miRNAs that are preferentially expressed in gonads. Gonadal selective miRNAs may play important roles in ovarian development, folliculogenesis and female fertility.
Figures

Similar articles
-
microRNA expression profiling of the developing mouse heart.Int J Mol Med. 2012 Nov;30(5):1095-104. doi: 10.3892/ijmm.2012.1092. Epub 2012 Aug 9. Int J Mol Med. 2012. PMID: 22895573
-
Identification of miRNAs during mouse postnatal ovarian development and superovulation.J Ovarian Res. 2015 Jul 8;8:44. doi: 10.1186/s13048-015-0170-2. J Ovarian Res. 2015. PMID: 26152307 Free PMC article.
-
Identification of the microRNA expression profile in the regenerative neonatal mouse heart by deep sequencing.Cell Biochem Biophys. 2014 Sep;70(1):635-42. doi: 10.1007/s12013-014-9967-7. Cell Biochem Biophys. 2014. PMID: 24756729
-
MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions.Int J Mol Sci. 2016 Oct 13;17(10):1712. doi: 10.3390/ijms17101712. Int J Mol Sci. 2016. PMID: 27754357 Free PMC article. Review.
-
The emerging role of microRNAs in fish ovary: A mini review.Gen Comp Endocrinol. 2021 Sep 15;311:113850. doi: 10.1016/j.ygcen.2021.113850. Epub 2021 Jul 7. Gen Comp Endocrinol. 2021. PMID: 34245767 Review.
Cited by
-
Transcriptome Analyses Identify Potential Key microRNAs and Their Target Genes Contributing to Ovarian Reserve.Int J Mol Sci. 2021 Oct 6;22(19):10819. doi: 10.3390/ijms221910819. Int J Mol Sci. 2021. PMID: 34639162 Free PMC article.
-
A microRNA signature of toxic extrasynaptic N-methyl-D-aspartate (NMDA) receptor signaling.Mol Brain. 2020 Jan 10;13(1):3. doi: 10.1186/s13041-020-0546-0. Mol Brain. 2020. PMID: 31924235 Free PMC article.
-
Genetic and epigenetic underpinnings of sex differences in the brain and in neurological and psychiatric disease susceptibility.Prog Brain Res. 2010;186:77-95. doi: 10.1016/B978-0-444-53630-3.00006-3. Prog Brain Res. 2010. PMID: 21094887 Free PMC article. Review.
-
MicroRNA Mediating Networks in Granulosa Cells Associated with Ovarian Follicular Development.Biomed Res Int. 2017;2017:4585213. doi: 10.1155/2017/4585213. Epub 2017 Feb 19. Biomed Res Int. 2017. PMID: 28316977 Free PMC article.
-
Interference of kallikrein 1b26 (klk1b26) translation by microRNA specifically expressed in female mouse submandibular glands: an additional mechanism for sexual dimorphism of klk1b26 protein in the glands.Biol Sex Differ. 2011 Nov 16;2:13. doi: 10.1186/2042-6410-2-13. Biol Sex Differ. 2011. PMID: 22085651 Free PMC article.
References
-
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi:10.1093/nar/25.17.3389. - DOI - PMC - PubMed
-
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi:10.1101/gad.1705308. - DOI - PMC - PubMed
-
- Ballow DJ, Xin Y, Choi Y, Pangas SA, Rajkovic A. Sohlh2 is a germ cell-specific bHLH transcription factor. Gene Expr Patterns. 2006;6:1014–1018. doi:10.1016/j.modgep.2006.04.007. - DOI - PubMed
-
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi:10.1016/j.devcel.2007.03.001. - DOI - PubMed
-
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi:10.1016/j.cell.2009.01.035. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

