Spliceosomal snRNA modifications and their function
- PMID: 20215871
- PMCID: PMC4154345
- DOI: 10.4161/rna.7.2.11207
Spliceosomal snRNA modifications and their function
Abstract
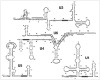
Spliceosomal snRNAs are extensively 2'-O-methylated and pseudouridylated. The modified nucleotides are relatively highly conserved across species, and are often clustered in regions of functional importance in pre-mRNA splicing. Over the past decade, the study of the mechanisms and functions of spliceosomal snRNA modifications has intensified. Two independent mechanisms behind these modifications, RNA-independent (protein-only) and RNA-dependent (RNA-guided), have been discovered. The role of spliceosomal snRNA modifications in snRNP biogenesis and spliceosome assembly has also been verified.
Figures



Similar articles
-
Functions and mechanisms of spliceosomal small nuclear RNA pseudouridylation.Wiley Interdiscip Rev RNA. 2011 Jul-Aug;2(4):571-81. doi: 10.1002/wrna.77. Epub 2011 Feb 18. Wiley Interdiscip Rev RNA. 2011. PMID: 21957045 Free PMC article. Review.
-
Pseudouridines in spliceosomal snRNAs.Protein Cell. 2011 Sep;2(9):712-25. doi: 10.1007/s13238-011-1087-1. Epub 2011 Oct 6. Protein Cell. 2011. PMID: 21976061 Free PMC article. Review.
-
Pseudouridine modification in Caenorhabditis elegans spliceosomal snRNAs: unique modifications are found in regions involved in snRNA-snRNA interactions.BMC Mol Biol. 2005 Oct 19;6:20. doi: 10.1186/1471-2199-6-20. BMC Mol Biol. 2005. PMID: 16236171 Free PMC article.
-
Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions.RNA. 1996 Sep;2(9):909-18. RNA. 1996. PMID: 8809017 Free PMC article.
-
Modified nucleotides at the 5' end of human U2 snRNA are required for spliceosomal E-complex formation.RNA. 2004 Dec;10(12):1925-33. doi: 10.1261/rna.7186504. Epub 2004 Nov 3. RNA. 2004. PMID: 15525712 Free PMC article.
Cited by
-
The epitranscriptome landscape of small noncoding RNAs in stem cells.Stem Cells. 2020 Oct 1;38(10):1216-1228. doi: 10.1002/stem.3233. Epub 2020 Jun 29. Stem Cells. 2020. PMID: 32598085 Free PMC article. Review.
-
Non-Coding RNAs in Breast Cancer: Intracellular and Intercellular Communication.Noncoding RNA. 2018 Dec 12;4(4):40. doi: 10.3390/ncrna4040040. Noncoding RNA. 2018. PMID: 30545127 Free PMC article. Review.
-
Site-Directed Spin Labeling of RNA with a Gem-Diethylisoindoline Spin Label: PELDOR, Relaxation, and Reduction Stability.Molecules. 2019 Dec 6;24(24):4482. doi: 10.3390/molecules24244482. Molecules. 2019. PMID: 31817785 Free PMC article.
-
The epitranscriptome of small non-coding RNAs.Noncoding RNA Res. 2021 Oct 26;6(4):167-173. doi: 10.1016/j.ncrna.2021.10.002. eCollection 2021 Dec. Noncoding RNA Res. 2021. PMID: 34820590 Free PMC article. Review.
-
Functions of RNA N6-methyladenosine modification in cancer progression.Mol Biol Rep. 2019 Apr;46(2):2567-2575. doi: 10.1007/s11033-019-04655-4. Epub 2019 Mar 25. Mol Biol Rep. 2019. PMID: 30911972 Review.
References
-
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell. 1998;92:315–26. - PubMed
-
- Yu YT, Scharl EC, Smith CM, Steitz JA. The RNA World. 2nd Cold Spring Harbor Laboratory Press; Cold Spring Harbor NY: 1999. pp. 487–524.
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. - PubMed
-
- Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr Opin Chem Biol. 2005;9:603–8. - PubMed
-
- Kramer A, Keller W, Appel B, Luhrmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984;38:299–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
