Spliceosomal snRNA modifications and their function
- PMID: 20215871
- PMCID: PMC4154345
- DOI: 10.4161/rna.7.2.11207
Spliceosomal snRNA modifications and their function
Abstract
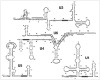
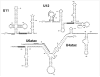
Spliceosomal snRNAs are extensively 2'-O-methylated and pseudouridylated. The modified nucleotides are relatively highly conserved across species, and are often clustered in regions of functional importance in pre-mRNA splicing. Over the past decade, the study of the mechanisms and functions of spliceosomal snRNA modifications has intensified. Two independent mechanisms behind these modifications, RNA-independent (protein-only) and RNA-dependent (RNA-guided), have been discovered. The role of spliceosomal snRNA modifications in snRNP biogenesis and spliceosome assembly has also been verified.
Figures



References
-
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell. 1998;92:315–26. - PubMed
-
- Yu YT, Scharl EC, Smith CM, Steitz JA. The RNA World. 2nd Cold Spring Harbor Laboratory Press; Cold Spring Harbor NY: 1999. pp. 487–524.
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. - PubMed
-
- Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr Opin Chem Biol. 2005;9:603–8. - PubMed
-
- Kramer A, Keller W, Appel B, Luhrmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984;38:299–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
