A microfluidic DNA computing processor for gene expression analysis and gene drug synthesis
- PMID: 20216967
- PMCID: PMC2835285
- DOI: 10.1063/1.3259628
A microfluidic DNA computing processor for gene expression analysis and gene drug synthesis
Abstract
Boolean logic performs a logical operation on one or more logic input and produces a single logic output. Here, we describe a microfluidic DNA computing processor performing Boolean logic operations for gene expression analysis and gene drug synthesis. Multiple cancer-related genes were used as input molecules. Their expression levels were identified by interacting with the computing related DNA strands, which were designed according to the sequences of cancer-related genes and the suicide gene. When all the expressions of the cancer-related genes fit in with the diagnostic criteria, positive diagnosis would be confirmed and then a complete suicide gene (gene drug) could be synthesized as an output molecule. Microfluidic chip was employed as an effective platform to realize the computing process by integrating multistep biochemical reactions involving hybridization, displacement, denaturalization, and ligation. By combining the specific design of the computing related molecules and the integrated functions of the microfluidics, the microfluidic DNA computing processor is able to analyze the multiple gene expressions simultaneously and realize the corresponding gene drug synthesis with simplicity and fast speed, which demonstrates the potential of this platform for DNA computing in biomedical applications.
Figures

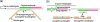
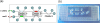



Similar articles
-
Programmable DNA-Based Boolean Logic Microfluidic Processing Unit.ACS Nano. 2021 Jul 27;15(7):11644-11654. doi: 10.1021/acsnano.1c02153. Epub 2021 Jul 7. ACS Nano. 2021. PMID: 34232017
-
An integrated microfluidic processor for single nucleotide polymorphism-based DNA computing.Lab Chip. 2005 Oct;5(10):1033-40. doi: 10.1039/b505840f. Epub 2005 Aug 11. Lab Chip. 2005. PMID: 16175257
-
Programmable DNA Nanoindicator-Based Platform for Large-Scale Square Root Logic Biocomputing.Small. 2019 Dec;15(49):e1903489. doi: 10.1002/smll.201903489. Epub 2019 Oct 29. Small. 2019. PMID: 31661189
-
Advancements in DNA computing: exploring DNA logic systems and their biomedical applications.J Mater Chem B. 2024 Oct 17;12(40):10134-10148. doi: 10.1039/d4tb00936c. J Mater Chem B. 2024. PMID: 39282799 Review.
-
Boolean Logic Gates Realized with Enzyme-catalyzed Reactions - Unusual Look at Usual Chemical Reactions.Chemphyschem. 2019 Jan 7;20(1):9-22. doi: 10.1002/cphc.201800900. Epub 2018 Nov 20. Chemphyschem. 2019. PMID: 30358034 Review.
Cited by
-
Multidimensional persistence in biomolecular data.J Comput Chem. 2015 Jul 30;36(20):1502-20. doi: 10.1002/jcc.23953. Epub 2015 May 30. J Comput Chem. 2015. PMID: 26032339 Free PMC article.
-
Fluorescence enhanced lab-on-a-chip patterned using a hybrid technique of femtosecond laser direct writing and anodized aluminum oxide porous nanostructuring.Nanoscale Adv. 2019 Jul 15;1(9):3474-3484. doi: 10.1039/c9na00352e. eCollection 2019 Sep 11. Nanoscale Adv. 2019. PMID: 36133573 Free PMC article.
-
Organosilane deposition for microfluidic applications.Biomicrofluidics. 2011 Sep;5(3):36501-365017. doi: 10.1063/1.3625605. Epub 2011 Aug 16. Biomicrofluidics. 2011. PMID: 22662048 Free PMC article.
-
Persistent homology analysis of protein structure, flexibility, and folding.Int J Numer Method Biomed Eng. 2014 Aug;30(8):814-44. doi: 10.1002/cnm.2655. Epub 2014 Jun 24. Int J Numer Method Biomed Eng. 2014. PMID: 24902720 Free PMC article.
References
-
- Johnson C. R., DNA Computing 4287, 360 (2006).10.1007/11925903_28 - DOI
LinkOut - more resources
Full Text Sources
