The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1
- PMID: 20219831
- PMCID: PMC2862436
- DOI: 10.1104/pp.109.150607
The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1
Abstract
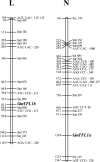
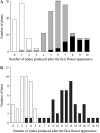
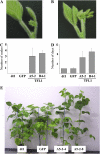
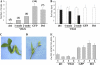
Classical genetic analysis has revealed that the determinate habit of soybean (Glycine max) is controlled by a recessive allele at the determinate stem (Dt1) locus. To dissect the molecular basis of the determinate habit, we isolated two orthologs of pea (Pisum sativum) TERMINAL FLOWER1a, GmTFL1a and GmTFL1b, from the soybean genome. Mapping analysis indicated that GmTFL1b is a candidate for Dt1. Despite their high amino acid identity, the two genes had different transcriptional profiles. GmTFL1b was expressed in the root and shoot apical meristems (SAMs), whereas GmTFL1a was mainly expressed in immature seed. The GmTFL1b transcript accumulated in the SAMs during early vegetative growth in both the determinate and indeterminate lines but thereafter was abruptly lost in the determinate line. Introduction of the genomic region of GmTFL1b from the indeterminate line complemented the stem growth habit in the determinate line: more nodes were produced, and flowering in the terminal raceme was delayed. The identity between Dt1 and GmTFL1b was also confirmed with a virus-induced gene silencing experiment. Taken together, our data suggest that Dt1 encodes the GmTFL1b protein and that the stem growth habit is determined by the variation of this gene. The dt1 allele may condition the determinate habit via the earlier loss in GmTFL1b expression concomitant with floral induction, although it functions normally under the noninductive phase of flowering. An association test of DNA polymorphisms with the stem growth habit among 16 cultivars suggested that a single amino acid substitution in exon 4 determines the fate of the SAM after floral induction.
Figures




References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 - PubMed
-
- Ablett GR, Beversdorf WD, Dirks VA. (1989) Performance and stability of indeterminate and determinate soybean in short-season environments. Crop Sci 29: 1428–1433
-
- Bernard RL. (1972) Two genes affecting stem termination in soybeans. Crop Sci 12: 235–239
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

