Genetic diversity of simian lentivirus in wild De Brazza's monkeys (Cercopithecus neglectus) in Equatorial Africa
- PMID: 20219893
- PMCID: PMC3052526
- DOI: 10.1099/vir.0.021048-0
Genetic diversity of simian lentivirus in wild De Brazza's monkeys (Cercopithecus neglectus) in Equatorial Africa
Abstract
De Brazza's monkeys (Cercopithecus neglectus) are non-human primates (NHP) living in Equatorial Africa from South Cameroon through the Congo-Basin to Uganda. As most of the NHP living in sub-Saharan Africa, they are naturally infected with their own simian lentivirus, SIVdeb. Previous studies confirmed this infection for De Brazza's from East Cameroon and Uganda. In this report, we studied the genetic diversity of SIVdeb in De Brazza's monkeys from different geographical areas in South Cameroon and from the Democratic Republic of Congo (DRC). SIVdeb strains from east, central and western equatorial Africa form a species-specific monophyletic lineage. Phylogeographic clustering was observed among SIVdeb strains from Cameroon, the DRC and Uganda, but also among primates from distinct areas in Cameroon. These observations suggest a longstanding virus-host co-evolution. SIVdeb prevalence is high in wild De Brazza's populations and thus represents a current risk for humans exposed to these primates in central Africa.
Figures

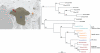
References
-
- Abascal, F., Zardoya, R. & Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. - PubMed
-
- Aghokeng, A. F. & Peeters, M. (2005). Simian immunodeficiency viruses (SIVs) in Africa. J Neurovirol 11, (Suppl. 1), 27–32. - PubMed
-
- Aghokeng, A. F., Bailes, E., Loul, S., Courgnaud, V., Mpoudi-Ngolle, E., Sharp, P. M., Delaporte, E. & Peeters, M. (2007). Full-length sequence analysis of SIVmus in wild populations of mustached monkeys (Cercopithecus cephus) from Cameroon provides evidence for two co-circulating SIVmus lineages. Virology 360, 407–418. - PMC - PubMed
-
- Aghokeng, A. F., Ayouba, A., Mpoudi-Ngole, E., Loul, S., Liegeois, F., Delaporte, E. & Peeters, M. (2010). Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infect Genet Evol 10, 386–396. - PMC - PubMed
-
- Bailes, E., Gao, F., Bibollet-Ruche, F., Courgnaud, V., Peeters, M., Marx, P. A., Hahn, B. H. & Sharp, P. M. (2003). Hybrid origin of SIV in chimpanzees. Science 300, 1713. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

