Influenza virus m2 ion channel protein is necessary for filamentous virion formation
- PMID: 20219914
- PMCID: PMC2863831
- DOI: 10.1128/JVI.00119-10
Influenza virus m2 ion channel protein is necessary for filamentous virion formation
Abstract
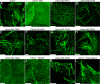
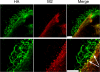
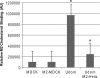
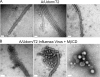
Influenza A virus buds from cells as spherical (approximately 100-nm diameter) and filamentous (approximately 100 nm x 2 to 20 microm) virions. Previous work has determined that the matrix protein (M1) confers the ability of the virus to form filaments; however, additional work has suggested that the influenza virus M2 integral membrane protein also plays a role in viral filament formation. In examining the role of the M2 protein in filament formation, we observed that the cytoplasmic tail of M2 contains several sites that are essential for filament formation. Additionally, whereas M2 is a nonraft protein, expression of other viral proteins in the context of influenza virus infection leads to the colocalization of M2 with sites of virus budding and lipid raft domains. We found that an amphipathic helix located within the M2 cytoplasmic tail is able to bind cholesterol, and we speculate that M2 cholesterol binding is essential for both filament formation and the stability of existing viral filaments.
Figures




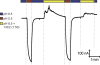
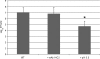
References
-
- Ada, G. L., B. T. Perry, and A. Abbot. 1958. Biological and physical properties of the Ryan strain of filamentous influenza virus. J. Gen. Microbiol. 19:23-39. - PubMed
-
- Balannik, V., R. A. Lamb, and L. H. Pinto. 2008. The oligomeric state of the active BM2 ion channel protein of influenza B virus. J. Biol. Chem. 283:4895-4904. - PubMed
-
- Bourmakina, S. V., and A. Garcia-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

