Regulation of Epstein-Barr virus OriP replication by poly(ADP-ribose) polymerase 1
- PMID: 20219917
- PMCID: PMC2863838
- DOI: 10.1128/JVI.02333-09
Regulation of Epstein-Barr virus OriP replication by poly(ADP-ribose) polymerase 1
Abstract
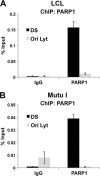
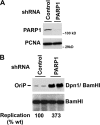
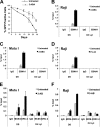
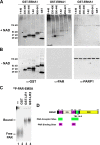
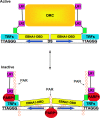
Poly(ADP-ribose) polymerase (PARP) is an abundant, chromatin-associated, NAD-dependent enzyme that functions in multiple chromosomal processes, including DNA replication and chromatin remodeling. The Epstein-Barr virus (EBV) origin of plasmid replication (OriP) is a dynamic genetic element that confers stable episome maintenance, DNA replication initiation, and chromatin organization functions. OriP function depends on the EBV-encoded origin binding protein EBNA1. We have previously shown that EBNA1 is subject to negative regulation by poly(ADP-ribosyl)ation (PARylation). We now show that PARP1 physically associates with OriP in latently EBV-infected B cells. Short hairpin RNA depletion of PARP1 enhances OriP replication activity and increases EBNA1, origin recognition complex 2 (ORC2), and minichromosome maintenance complex (MCM) association with OriP. Pharmacological inhibitors of PARP1 enhance OriP plasmid maintenance and increase EBNA1, ORC2, and MCM3 occupancy at OriP. PARylation in vitro inhibits ORC2 recruitment and remodels telomere repeat factor (TRF) binding at the dyad symmetry (DS) element of OriP. Purified PARP1 can ribosylate EBNA1 at multiple sites throughout its amino terminus but not in the carboxy-terminal DNA binding domain. We also show that EBNA1 linking regions (LR1 and LR2) can bind directly to oligomers of PAR. We propose that PARP1-dependent PARylation of EBNA1 and adjacently bound TRF2 induces structural changes at the DS element that reduce EBNA1 DNA binding affinity and functional recruitment of ORC.
Figures
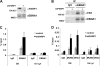


Similar articles
-
Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication.J Virol. 2019 Sep 30;93(20):e00487-19. doi: 10.1128/JVI.00487-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31142669 Free PMC article.
-
Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1.J Virol. 2005 Apr;79(8):4640-50. doi: 10.1128/JVI.79.8.4640-4650.2005. J Virol. 2005. PMID: 15795250 Free PMC article.
-
Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication.Mol Cell. 2002 Mar;9(3):493-503. doi: 10.1016/s1097-2765(02)00476-8. Mol Cell. 2002. PMID: 11931758
-
Replication licensing of the EBV oriP minichromosome.Curr Top Microbiol Immunol. 2001;258:13-33. doi: 10.1007/978-3-642-56515-1_2. Curr Top Microbiol Immunol. 2001. PMID: 11443858 Review.
-
EBNA1 and host factors in Epstein-Barr virus latent DNA replication.Curr Opin Virol. 2012 Dec;2(6):733-9. doi: 10.1016/j.coviro.2012.09.005. Epub 2012 Sep 30. Curr Opin Virol. 2012. PMID: 23031715 Review.
Cited by
-
Bookmarking promoters in mitotic chromatin: poly(ADP-ribose)polymerase-1 as an epigenetic mark.Nucleic Acids Res. 2014 Jun;42(11):7028-38. doi: 10.1093/nar/gku415. Epub 2014 May 26. Nucleic Acids Res. 2014. PMID: 24861619 Free PMC article.
-
SIRT1 deacetylates TopBP1 and modulates intra-S-phase checkpoint and DNA replication origin firing.Int J Biol Sci. 2014 Nov 26;10(10):1193-202. doi: 10.7150/ijbs.11066. eCollection 2014. Int J Biol Sci. 2014. PMID: 25516717 Free PMC article.
-
The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition.PLoS Genet. 2015 May 22;11(5):e1005252. doi: 10.1371/journal.pgen.1005252. eCollection 2015 May. PLoS Genet. 2015. PMID: 26001115 Free PMC article.
-
Epigenetic regulation of EBV and KSHV latency.Curr Opin Virol. 2013 Jun;3(3):251-9. doi: 10.1016/j.coviro.2013.03.004. Epub 2013 Apr 16. Curr Opin Virol. 2013. PMID: 23601957 Free PMC article. Review.
-
Viral Macrodomains: Unique Mediators of Viral Replication and Pathogenesis.Trends Microbiol. 2018 Jul;26(7):598-610. doi: 10.1016/j.tim.2017.11.011. Trends Microbiol. 2018. PMID: 29268982 Free PMC article. Review.
References
-
- Ahel, I., D. Ahel, T. Matsusaka, A. J. Clark, J. Pines, S. J. Boulton, and S. C. West. 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451:81-85. - PubMed
-
- Allinson, S. L., I. I. Dianova, and G. L. Dianov. 2003. Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim. Pol. 50:169-179. - PubMed
-
- Ame, J. C., C. Spenlehauer, and G. De Murcia. 2004. The PARP superfamily. Bioessays 26:882-893. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

