Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses
- PMID: 20219926
- PMCID: PMC2863824
- DOI: 10.1128/JVI.00057-10
Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses
Abstract
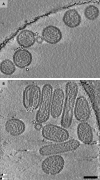
Hantaviruses (family Bunyaviridae) are rodent-borne emerging viruses that cause a serious, worldwide threat to human health. Hantavirus diseases include hemorrhagic fever with renal syndrome and hantavirus cardiopulmonary syndrome. Virions are enveloped and contain a tripartite single-stranded negative-sense RNA genome. Two types of glycoproteins, G(N) and G(C), are embedded in the viral membrane and form protrusions, or "spikes." The membrane encloses a ribonucleoprotein core, which consists of the RNA segments, the nucleocapsid protein, and the RNA-dependent RNA polymerase. Detailed information on hantavirus virion structure and glycoprotein spike composition is scarce. Here, we have studied the structures of Tula hantavirus virions using electron cryomicroscopy and tomography. Three-dimensional density maps show how the hantavirus surface glycoproteins, membrane, and ribonucleoprotein are organized. The structure of the G(N)-G(C) spike complex was solved to 3.6-nm resolution by averaging tomographic subvolumes. Each spike complex is a square-shaped assembly with 4-fold symmetry. Spike complexes formed ordered patches on the viral membrane by means of specific lateral interactions. These interactions may be sufficient for creating membrane curvature during virus budding. In conclusion, the structure and assembly principles of Tula hantavirus exemplify a unique assembly paradigm for enveloped viruses.
Figures




References
-
- Adrian, M., J. Dubochet, J. Lepault, and A. W. McDowall. 1984. Cryo-electron microscopy of viruses. Nature 308:32-36. - PubMed
-
- Deyde, V. M., A. A. Rizvanov, J. Chase, E. W. Otteson, and S. C. St. Jeor. 2005. Interactions and trafficking of Andes and Sin Nombre Hantavirus glycoproteins G1 and G2. Virology 331:307-315. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

