A novel molecular mechanism of dual resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors
- PMID: 20219933
- PMCID: PMC2863829
- DOI: 10.1128/JVI.01545-09
A novel molecular mechanism of dual resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors
Abstract
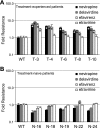
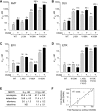
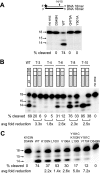
Recently, mutations in the connection subdomain (CN) and RNase H domain of HIV-1 reverse transcriptase (RT) were observed to exhibit dual resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors (NRTIs and NNRTIs). To elucidate the mechanism by which CN and RH mutations confer resistance to NNRTIs, we hypothesized that these mutations reduce RNase H cleavage and provide more time for the NNRTI to dissociate from the RT, resulting in the resumption of DNA synthesis and enhanced NNRTI resistance. We observed that the effect of the reduction in RNase H cleavage on NNRTI resistance is dependent upon the affinity of each NNRTI to the RT and further influenced by the presence of NNRTI-binding pocket (BP) mutants. D549N, Q475A, and Y501A mutants, which reduce RNase H cleavage, enhance resistance to nevirapine (NVP) and delavirdine (DLV), but not to efavirenz (EFV) and etravirine (ETR), consistent with their increase in affinity for RT. Combining the D549N mutant with NNRTI BP mutants further increases NNRTI resistance from 3- to 30-fold, supporting the role of NNRTI-RT affinity in our NNRTI resistance model. We also demonstrated that CNs from treatment-experienced patients, previously reported to enhance NRTI resistance, also reduce RNase H cleavage and enhance NNRTI resistance in the context of the patient RT pol domain or a wild-type pol domain. Together, these results confirm key predictions of our NNRTI resistance model and provide support for a unifying mechanism by which CN and RH mutations can exhibit dual NRTI and NNRTI resistance.
Figures


Similar articles
-
Combinations of mutations in the connection domain of human immunodeficiency virus type 1 reverse transcriptase: assessing the impact on nucleoside and nonnucleoside reverse transcriptase inhibitor resistance.Antimicrob Agents Chemother. 2010 May;54(5):1973-80. doi: 10.1128/AAC.00870-09. Epub 2010 Mar 1. Antimicrob Agents Chemother. 2010. PMID: 20194692 Free PMC article.
-
HIV-1 reverse transcriptase connection subdomain mutations involved in resistance to approved non-nucleoside inhibitors.Antiviral Res. 2011 Nov;92(2):139-49. doi: 10.1016/j.antiviral.2011.08.020. Epub 2011 Aug 28. Antiviral Res. 2011. PMID: 21896288 Review.
-
Altered viral fitness and drug susceptibility in HIV-1 carrying mutations that confer resistance to nonnucleoside reverse transcriptase and integrase strand transfer inhibitors.J Virol. 2014 Aug;88(16):9268-76. doi: 10.1128/JVI.00695-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899199 Free PMC article.
-
The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5'-end- and DNA 3'-end-directed RNase H activities.J Virol. 1999 Jul;73(7):5803-13. doi: 10.1128/JVI.73.7.5803-5813.1999. J Virol. 1999. PMID: 10364332 Free PMC article.
-
Current status of the non-nucleoside reverse transcriptase inhibitors of human immunodeficiency virus type 1.Curr Top Med Chem. 2004;4(9):921-44. doi: 10.2174/1568026043388420. Curr Top Med Chem. 2004. PMID: 15134549 Review.
Cited by
-
Phenotypic characterization of drug resistance-associated mutations in HIV-1 RT connection and RNase H domains and their correlation with thymidine analogue mutations.J Antimicrob Chemother. 2011 Apr;66(4):702-8. doi: 10.1093/jac/dkr005. Epub 2011 Jan 26. J Antimicrob Chemother. 2011. PMID: 21393163 Free PMC article.
-
Noncanonical HIV drug resistance mutations: need to close existing gaps.AIDS. 2025 Jun 1;39(7):781-787. doi: 10.1097/QAD.0000000000004170. Epub 2025 Mar 17. AIDS. 2025. PMID: 40053490 Free PMC article. Review.
-
The connection domain mutation N348I in HIV-1 reverse transcriptase enhances resistance to etravirine and rilpivirine but restricts the emergence of the E138K resistance mutation by diminishing viral replication capacity.J Virol. 2014 Feb;88(3):1536-47. doi: 10.1128/JVI.02904-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227862 Free PMC article.
-
Impact of the N348I mutation in HIV-1 reverse transcriptase on nonnucleoside reverse transcriptase inhibitor resistance in non-subtype B HIV-1.Antimicrob Agents Chemother. 2011 Apr;55(4):1806-9. doi: 10.1128/AAC.01197-10. Epub 2011 Jan 31. Antimicrob Agents Chemother. 2011. PMID: 21282419 Free PMC article.
-
A polymorphism at position 400 in the connection subdomain of HIV-1 reverse transcriptase affects sensitivity to NNRTIs and RNaseH activity.PLoS One. 2013 Oct 2;8(10):e74078. doi: 10.1371/journal.pone.0074078. eCollection 2013. PLoS One. 2013. PMID: 24098331 Free PMC article.
References
-
- Andries, K., H. Azijn, T. Thielemans, D. Ludovici, M. Kukla, J. Heeres, P. Janssen, B. De Corte, J. Vingerhoets, R. Pauwels, and M. P. de Bethune. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680-4686. - PMC - PubMed
-
- Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. - PMC - PubMed
-
- Archer, R. H., M. Wisniewski, R. A. Bambara, and L. M. Demeter. 2001. The Y181C mutant of HIV-1 reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors alters the size distribution of RNase H cleavages. Biochemistry 40:4087-4095. - PubMed
-
- Baldanti, F., S. Paolucci, G. Maga, N. Labo, U. Hubscher, A. Y. Skoblov, L. Victorova, S. Spadari, L. Minoli, and G. Gerna. 2003. Nevirapine-selected mutations Y181I/C of HIV-1 reverse transcriptase confer cross-resistance to stavudine. AIDS 17:1568-1570. - PubMed
-
- Blanca, G., F. Baldanti, S. Paolucci, A. Y. Skoblov, L. Victorova, U. Hubscher, G. Gerna, S. Spadari, and G. Maga. 2003. Nevirapine resistance mutation at codon 181 of the HIV-1 reverse transcriptase confers stavudine resistance by increasing nucleotide substrate discrimination and phosphorolytic activity. J. Biol. Chem. 278:15469-15472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

