Effect of ATP binding and hydrolysis on dynamics of canine parvovirus NS1
- PMID: 20219935
- PMCID: PMC2863850
- DOI: 10.1128/JVI.02221-09
Effect of ATP binding and hydrolysis on dynamics of canine parvovirus NS1
Abstract
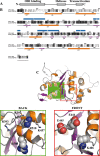
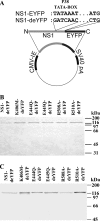
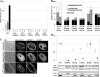
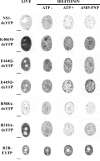
The replication protein NS1 is essential for genome replication and protein production in parvoviral infection. Many of its functions, including recognition and site-specific nicking of the viral genome, helicase activity, and transactivation of the viral capsid promoter, are dependent on ATP. An ATP-binding pocket resides in the middle of the modular NS1 protein in a superfamily 3 helicase domain. Here we have identified key ATP-binding amino acid residues in canine parvovirus (CPV) NS1 protein and mutated amino acids from the conserved A motif (K406), B motif (E444 and E445), and positively charged region (R508 and R510). All mutations prevented the formation of infectious viruses. When provided in trans, all except the R508A mutation reduced infectivity in a dominant-negative manner, possibly by hindering genome replication. These results suggest that the conserved R510 residue, but not R508, is the arginine finger sensory element of CPV NS1. Moreover, fluorescence recovery after photobleaching (FRAP), complemented by computer simulations, was used to assess the binding properties of mutated fluorescent fusion proteins. These experiments identified ATP-dependent and -independent binding modes for NS1 in living cells. Only the K406M mutant had a single binding site, which was concluded to indicate ATP-independent binding. Furthermore, our data suggest that DNA binding of NS1 is dependent on its ability to both bind and hydrolyze ATP.
Figures

Similar articles
-
Structure and function of the parvoviral NS1 protein: a review.Virus Genes. 2023 Apr;59(2):195-203. doi: 10.1007/s11262-022-01944-2. Epub 2022 Oct 17. Virus Genes. 2023. PMID: 36253516 Review.
-
Nonstructural protein-2 and the replication of canine parvovirus.Virology. 1998 Jan 20;240(2):273-81. doi: 10.1006/viro.1997.8946. Virology. 1998. PMID: 9454701
-
Mutations in DNA binding and transactivation domains affect the dynamics of parvovirus NS1 protein.J Virol. 2013 Nov;87(21):11762-74. doi: 10.1128/JVI.01678-13. Epub 2013 Aug 28. J Virol. 2013. PMID: 23986577 Free PMC article.
-
Biochemical characterization of Periplaneta fuliginosa densovirus non-structural protein NS1.Biochem Biophys Res Commun. 2006 Apr 21;342(4):1188-96. doi: 10.1016/j.bbrc.2006.02.053. Epub 2006 Feb 20. Biochem Biophys Res Commun. 2006. PMID: 16516861
-
Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid.Virology. 1995 Aug 1;211(1):123-32. doi: 10.1006/viro.1995.1385. Virology. 1995. PMID: 7645206
Cited by
-
Canine Parvovirus and Its Non-Structural Gene 1 as Oncolytic Agents: Mechanism of Action and Induction of Anti-Tumor Immune Response.Front Oncol. 2021 May 3;11:648873. doi: 10.3389/fonc.2021.648873. eCollection 2021. Front Oncol. 2021. PMID: 34012915 Free PMC article. Review.
-
Structure and function of the parvoviral NS1 protein: a review.Virus Genes. 2023 Apr;59(2):195-203. doi: 10.1007/s11262-022-01944-2. Epub 2022 Oct 17. Virus Genes. 2023. PMID: 36253516 Review.
-
Novel mutation N588 residue in the NS1 protein of feline parvovirus greatly augments viral replication.J Virol. 2024 May 14;98(5):e0009324. doi: 10.1128/jvi.00093-24. Epub 2024 Apr 9. J Virol. 2024. PMID: 38591899 Free PMC article.
-
Promoter-Targeted Histone Acetylation of Chromatinized Parvoviral Genome Is Essential for the Progress of Infection.J Virol. 2016 Mar 28;90(8):4059-4066. doi: 10.1128/JVI.03160-15. Print 2016 Apr. J Virol. 2016. PMID: 26842481 Free PMC article.
-
Phylogenetic analysis and molecular structure of NS1 proteins of porcine parvovirus 5 isolates from Mexico.Arch Virol. 2025 Jan 24;170(2):40. doi: 10.1007/s00705-024-06182-5. Arch Virol. 2025. PMID: 39856382 Free PMC article.
References
-
- Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42.
-
- Barton, G. J. 1993. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 6:37-40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

