Histone methylation regulates memory formation
- PMID: 20219993
- PMCID: PMC2859898
- DOI: 10.1523/JNEUROSCI.3732-09.2010
Histone methylation regulates memory formation
Abstract
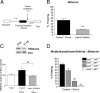
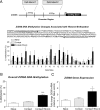
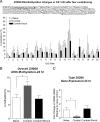
It has been established that regulation of chromatin structure through post-translational modification of histone proteins, primarily histone H3 phosphorylation and acetylation, is an important early step in the induction of synaptic plasticity and formation of long-term memory. In this study, we investigated the contribution of another histone modification, histone methylation, to memory formation in the adult hippocampus. We found that trimethylation of histone H3 at lysine 4 (H3K4), an active mark for transcription, is upregulated in hippocampus 1 h following contextual fear conditioning. In addition, we found that dimethylation of histone H3 at lysine 9 (H3K9), a molecular mark associated with transcriptional silencing, is increased 1 h after fear conditioning and decreased 24 h after context exposure alone and contextual fear conditioning. Trimethylated H3K4 levels returned to baseline levels at 24 h. We also found that mice deficient in the H3K4-specific histone methyltransferase, Mll, displayed deficits in contextual fear conditioning relative to wild-type animals. This suggests that histone methylation is required for proper long-term consolidation of contextual fear memories. Interestingly, inhibition of histone deacetylases (HDACs) with sodium butyrate (NaB) resulted in increased H3K4 trimethylation and decreased H3K9 dimethylation in hippocampus following contextual fear conditioning. Correspondingly, we found that fear learning triggered increases in H3K4 trimethylation at specific gene promoter regions (Zif268 and bdnf) with altered DNA methylation and MeCP2 DNA binding. Zif268 DNA methylation levels returned to baseline at 24 h. Together, these data demonstrate that histone methylation is actively regulated in the hippocampus and facilitates long-term memory formation.
Figures





Similar articles
-
G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation.J Neurosci. 2012 Apr 18;32(16):5440-53. doi: 10.1523/JNEUROSCI.0147-12.2012. J Neurosci. 2012. PMID: 22514307 Free PMC article.
-
Histone acetylation rescues contextual fear conditioning in nNOS KO mice and accelerates extinction of cued fear conditioning in wild type mice.Neurobiol Learn Mem. 2012 May;97(4):409-17. doi: 10.1016/j.nlm.2012.03.005. Epub 2012 Mar 20. Neurobiol Learn Mem. 2012. PMID: 22452925 Free PMC article.
-
Epigenetic modulation of Homer1a transcription regulation in amygdala and hippocampus with pavlovian fear conditioning.J Neurosci. 2012 Mar 28;32(13):4651-9. doi: 10.1523/JNEUROSCI.3308-11.2012. J Neurosci. 2012. PMID: 22457511 Free PMC article.
-
Epigenetic landscape of amphetamine and methamphetamine addiction in rodents.Epigenetics. 2015;10(7):574-80. doi: 10.1080/15592294.2015.1055441. Epigenetics. 2015. PMID: 26023847 Free PMC article. Review.
-
Histone H3 lysine K4 methylation and its role in learning and memory.Epigenetics Chromatin. 2019 Jan 7;12(1):7. doi: 10.1186/s13072-018-0251-8. Epigenetics Chromatin. 2019. PMID: 30616667 Free PMC article. Review.
Cited by
-
Nuclear factor κB-dependent histone acetylation is specifically involved in persistent forms of memory.J Neurosci. 2013 Apr 24;33(17):7603-14. doi: 10.1523/JNEUROSCI.4181-12.2013. J Neurosci. 2013. PMID: 23616565 Free PMC article.
-
Epigenetic mechanisms underlying learning and the inheritance of learned behaviors.Trends Neurosci. 2015 Feb;38(2):96-107. doi: 10.1016/j.tins.2014.12.003. Epub 2014 Dec 24. Trends Neurosci. 2015. PMID: 25544352 Free PMC article. Review.
-
Neuroprotective actions of perinatal choline nutrition.Clin Chem Lab Med. 2013 Mar 1;51(3):591-9. doi: 10.1515/cclm-2012-0635. Clin Chem Lab Med. 2013. PMID: 23314544 Free PMC article. Review.
-
Exploring epigenetic regulation of fear memory and biomarkers associated with post-traumatic stress disorder.Front Psychiatry. 2013 Jul 1;4:62. doi: 10.3389/fpsyt.2013.00062. eCollection 2013. Front Psychiatry. 2013. PMID: 23847551 Free PMC article.
-
Detection of histone acetylation levels in the dorsal hippocampus reveals early tagging on specific residues of H2B and H4 histones in response to learning.PLoS One. 2013;8(3):e57816. doi: 10.1371/journal.pone.0057816. Epub 2013 Mar 4. PLoS One. 2013. Retraction in: PLoS One. 2023 Dec 22;18(12):e0296474. doi: 10.1371/journal.pone.0296474. PMID: 23469244 Free PMC article. Retracted.
References
-
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. - PubMed
-
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. - PubMed
-
- Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG031722/AG/NIA NIH HHS/United States
- NS37444/NS/NINDS NIH HHS/United States
- P30 NS057098/NS/NINDS NIH HHS/United States
- NS013546/NS/NINDS NIH HHS/United States
- R01 NS013546/NS/NINDS NIH HHS/United States
- R01 AG031722/AG/NIA NIH HHS/United States
- NS057098/NS/NINDS NIH HHS/United States
- R01 MH057014/MH/NIMH NIH HHS/United States
- P01 NS037444/NS/NINDS NIH HHS/United States
- MH082106/MH/NIMH NIH HHS/United States
- NS048811/NS/NINDS NIH HHS/United States
- MH57014/MH/NIMH NIH HHS/United States
- K99 MH082106/MH/NIMH NIH HHS/United States
- R01 MH097909/MH/NIMH NIH HHS/United States
- F32 NS048811/NS/NINDS NIH HHS/United States
- R00 MH082106/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
