The nature of peptides presented by an HLA class I low expression allele
- PMID: 20220067
- PMCID: PMC2913087
- DOI: 10.3324/haematol.2009.016089
The nature of peptides presented by an HLA class I low expression allele
Abstract
Background: The functional integrity of human leukocyte antigen low expression variants is a prerequisite for considering them as essential in the matching process of hematopoietic stem cell donors and recipients to diminish the risk of serious complications such as graft-versus-host disease or graft rejection. The HLA-A*3014L variant has a disulfide bridge missing in the alpha2 domain which could affect peptide binding and presentation to T cells.
Design and methods: HLA-A*3014L and HLA-A*3001 were expressed as truncated variants and peptides were eluted and subjected to pool sequencing by Edman degradation as well as to single-peptide sequencing by mass spectrometry. Quantitative analysis of binding peptides presented in vivo was performed by a flow cytometric peptide-binding assay using HLA-A*3001 and HLA-A*3014L-expressing B-LCLs.
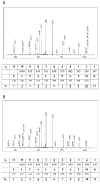
Results: The truncated HLA-A*3014L protein was secreted in the supernatant and it was possible to elute and sequence peptides. Sequence analysis of these eluted peptides revealed no relevant differences to the peptide motif of HLA-A*3001, indicating that the Cys164Ser substitution does not substantially alter the spectrum of presented peptides. Strong binding of one of the shared in vivo identified HLA-A*3001/3014L ligands was confirmed in the peptide-binding assay.
Conclusions: This study is the first to demonstrate that HLA low expression variants are able to present peptides and, thus, can be considered as functionally active. When comparing peptide motifs, it is likely that HLA-A*3014L and HLA-A*3001 represent a permissive mismatch with low allogenicity in hematopoietic stem cell transplantation. These results indicate that surface expression, as well as peptide-binding data of HLA variants with similar disulfide bridge variations (e.g. HLA-A*3211Q) need to be considered as functionally active in an allogeneic hematopoietic stem cell transplantation setting as long as the opposite has not been shown. Otherwise a relevant but not considered HLA mismatch could result in a severe allogeneic T-cell response and graft-versus-host disease.
Figures

Similar articles
-
Discrimination of HLA null and low expression alleles by cytokine-induced secretion of recombinant soluble HLA.Mol Immunol. 2009 Apr;46(7):1451-7. doi: 10.1016/j.molimm.2008.12.011. Epub 2009 Feb 4. Mol Immunol. 2009. PMID: 19195705
-
Definition of peptide binding motifs amongst the HLA-A*30 allelic group.Tissue Antigens. 2000 Jul;56(1):10-8. doi: 10.1034/j.1399-0039.2000.560102.x. Tissue Antigens. 2000. PMID: 10958351
-
Mismatches outside exons 2 and 3 do not alter the peptide motif of the allele group B*44:02P.Hum Immunol. 2011 Nov;72(11):1039-44. doi: 10.1016/j.humimm.2011.08.004. Epub 2011 Aug 10. Hum Immunol. 2011. PMID: 21872626
-
Class I MHC-peptide interactions: structural requirements and functional implications.Cancer Surv. 1995;22:37-49. Cancer Surv. 1995. PMID: 7536628 Review.
-
HLA-E: a novel player for histocompatibility.J Immunol Res. 2014;2014:352160. doi: 10.1155/2014/352160. Epub 2014 Oct 20. J Immunol Res. 2014. PMID: 25401109 Free PMC article. Review.
Cited by
-
Immunoinformatics study to search epitopes of spike glycoprotein from SARS-CoV-2 as potential vaccine.J Biomol Struct Dyn. 2021 Aug;39(13):4878-4892. doi: 10.1080/07391102.2020.1780944. Epub 2020 Jun 25. J Biomol Struct Dyn. 2021. PMID: 32583729 Free PMC article.
-
Position 45 influences the peptide binding motif of HLA-B*44:08.Immunogenetics. 2012 Mar;64(3):245-9. doi: 10.1007/s00251-011-0583-z. Epub 2011 Oct 19. Immunogenetics. 2012. PMID: 22009320
-
Personalized adoptive immunotherapy for patients with EBV-associated tumors and complications: Evaluation of novel naturally processed and presented EBV-derived T-cell epitopes.Oncotarget. 2017 Dec 21;9(4):4737-4757. doi: 10.18632/oncotarget.23531. eCollection 2018 Jan 12. Oncotarget. 2017. PMID: 29435138 Free PMC article.
-
Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.Front Immunol. 2019 Jul 23;10:1709. doi: 10.3389/fimmu.2019.01709. eCollection 2019. Front Immunol. 2019. PMID: 31396224 Free PMC article.
References
-
- Natarajan K, Li H, Mariuzza RA, Margulies DH. MHC class I molecules, structure and function. Rev Immunogenet. 1999;1(1):32–46. - PubMed
-
- Bade-Doeding C, Elsner HA, Eiz-Vesper B, Seltsam A, Holtkamp U, Blasczyk R. A single amino-acid polymorphism in pocket A of HLA-A*6602 alters the auxiliary anchors compared with HLA-A*6601 ligands. Immunogenetics. 2004;56(2):83–8. - PubMed
-
- Prilliman KR, Jackson KW, Lindsey M, Wang J, Crawford D, Hildebrand WH. HLA-B15 peptide ligands are preferentially anchored at their C termini. J Immunol. 1999;162(12):7277–84. - PubMed
-
- Hirv K, Pannicke U, Mytilineos J, Schwarz K. Disulfide bridge disruption in the alpha2 domain of the HLA class I molecule leads to low expression of the corresponding antigen. Hum Immunol. 2006;67(8):589–96. - PubMed
-
- Warburton RJ, Matsui M, Rowland-Jones SL, Gammon MC, Katzenstein GE, Wei T, et al. Mutation of the alpha 2 domain disulfide bridge of the class I molecule HLA-A*0201. Effect on maturation and peptide presentation. Hum Immunol. 1994;39(4):261–71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

