Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice
- PMID: 20220098
- PMCID: PMC2851884
- DOI: 10.1073/pnas.0911965107
Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice
Abstract
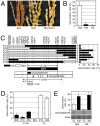
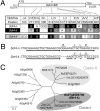
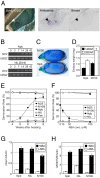
Seed dormancy provides a strategy for flowering plants to survive adverse natural conditions. It is also an important agronomic trait affecting grain yield, quality, and processing performance. We cloned a rice quantitative trait locus, Sdr4, which contributes substantially to differences in seed dormancy between japonica (Nipponbare) and indica (Kasalath) cultivars. Sdr4 expression is positively regulated by OsVP1, a global regulator of seed maturation, and in turn positively regulates potential regulators of seed dormancy and represses the expression of postgerminative genes, suggesting that Sdr4 acts as an intermediate regulator of dormancy in the seed maturation program. Japonica cultivars have only the Nipponbare allele (Sdr4-n), which endows reduced dormancy, whereas both the Kasalath allele (Srd4-k) and Sdr4-n are widely distributed in the indica group, indicating prevalent introgression. Srd4-k also is found in the wild ancestor Oryza rufipogon, whereas Sdr4-n appears to have been produced through at least two mutation events from the closest O. rufipogon allele among the accessions examined. These results are discussed with respect to possible selection of the allele during the domestication process.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Sdr4 dominates pre-harvest sprouting and facilitates adaptation to local climatic condition in Asian cultivated rice.J Integr Plant Biol. 2022 Jun;64(6):1246-1263. doi: 10.1111/jipb.13266. Epub 2022 May 31. J Integr Plant Biol. 2022. PMID: 35442537
-
Estimation of loci involved in non-shattering of seeds in early rice domestication.Genetica. 2017 Apr;145(2):201-207. doi: 10.1007/s10709-017-9958-x. Epub 2017 Feb 25. Genetica. 2017. PMID: 28238052
-
Transcriptome analysis of knockout mutants of rice seed dormancy gene OsVP1 and Sdr4.Plant Cell Rep. 2023 Feb;42(2):309-319. doi: 10.1007/s00299-022-02958-8. Epub 2022 Nov 29. Plant Cell Rep. 2023. PMID: 36445461
-
[Major domestication traits in Asian rice].Yi Chuan. 2012 Nov;34(11):1379-89. doi: 10.3724/sp.j.1005.2012.01379. Yi Chuan. 2012. PMID: 23208135 Review. Chinese.
-
The complex history of the domestication of rice.Ann Bot. 2007 Nov;100(5):951-7. doi: 10.1093/aob/mcm128. Epub 2007 Jul 6. Ann Bot. 2007. PMID: 17617555 Free PMC article. Review.
Cited by
-
A mediator of OsbZIP46 deactivation and degradation negatively regulates seed dormancy in rice.Nat Commun. 2024 Feb 7;15(1):1134. doi: 10.1038/s41467-024-45402-z. Nat Commun. 2024. PMID: 38326370 Free PMC article.
-
Screening of NIAS World Rice Core Collection for Seeds with Long Longevity as Useful Potential Breeding Materials Focusing on the Stability of Embryonic RNAs.Plants (Basel). 2024 Jul 6;13(13):1869. doi: 10.3390/plants13131869. Plants (Basel). 2024. PMID: 38999709 Free PMC article.
-
Genome-wide characterization of SDR gene family and its potential role in seed dormancy of Brassica napus L.BMC Plant Biol. 2024 Jan 2;24(1):21. doi: 10.1186/s12870-023-04700-2. BMC Plant Biol. 2024. PMID: 38166550 Free PMC article.
-
Genetic dissection of domestication-related traits in soybean through genotyping-by-sequencing of two interspecific mapping populations.Theor Appl Genet. 2019 Apr;132(4):1195-1209. doi: 10.1007/s00122-018-3272-6. Epub 2019 Jan 3. Theor Appl Genet. 2019. PMID: 30607438
-
Editorial: Seed Dormancy, Germination, and Pre-harvest Sprouting.Front Plant Sci. 2018 Nov 30;9:1783. doi: 10.3389/fpls.2018.01783. eCollection 2018. Front Plant Sci. 2018. PMID: 30555506 Free PMC article. No abstract available.
References
-
- Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23:578–587. - PubMed
-
- Izawa T, Konishi S, Shomura A, Yano M. DNA changes tell us about rice domestication. Curr Opin Plant Biol. 2009;12:185–192. - PubMed
-
- Hilhorst HWM. In: Seed Development, Dormancy and Germination. Bradford KJ, Nonogaki H, editors. Sheffield, UK: Blackwell; 2007. pp. 50–71.
-
- McCarty DR, et al. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

