Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B
- PMID: 20220129
- PMCID: PMC2863241
- DOI: 10.1074/jbc.M110.111914
Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B
Abstract
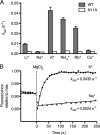
FeoB is a prokaryotic membrane protein responsible for the import of ferrous iron (Fe(2+)). A defining feature of FeoB is that it includes an N-terminal 30-kDa soluble domain with GTPase activity, which is required for iron transport. However, the low intrinsic GTP hydrolysis rate of this domain appears to be too slow for FeoB either to function as a channel or to possess an active Fe(2+) membrane transport mechanism. Here, we present crystal structures of the soluble domain of FeoB from Streptococcus thermophilus in complex with GDP and with the GTP analogue derivative 2'-(or -3')-O-(N-methylanthraniloyl)-beta,gamma-imidoguanosine 5'-triphosphate (mant-GMPPNP). Unlike recent structures of the G protein domain, the mant-GMPPNP-bound structure shows clearly resolved, active conformations of the critical Switch motifs. Importantly, biochemical analyses demonstrate that the GTPase activity of FeoB is activated by K(+), which leads to a 20-fold acceleration in its hydrolysis rate. Analysis of the structure identified a conserved asparagine residue likely to be involved in K(+) coordination, and mutation of this residue abolished K(+)-dependent activation. We suggest that this, together with a second asparagine residue that we show is critical for the structure of the Switch I loop, allows the prediction of K(+)-dependent activation in G proteins. In addition, the accelerated hydrolysis rate opens up the possibility that FeoB might indeed function as an active transporter.
Figures


References
-
- Cachero T. G., Morielli A. D., Peralta E. G. (1998) Cell 93, 1077–1085 - PubMed
-
- Finlin B. S., Correll R. N., Pang C., Crump S. M., Satin J., Andres D. A. (2006) J. Biol. Chem. 281, 23557–23566 - PubMed
-
- Schaafsma D., Roscioni S. S., Meurs H., Schmidt M. (2008) Cell Signal. 20, 1705–1714 - PubMed
-
- Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

