Mitochondrial and nuclear genomic responses to loss of LRPPRC expression
- PMID: 20220140
- PMCID: PMC2859537
- DOI: 10.1074/jbc.M109.098400
Mitochondrial and nuclear genomic responses to loss of LRPPRC expression
Erratum in
-
Correction: Mitochondrial and nuclear genomic responses to loss of LRPPRC expression.J Biol Chem. 2020 Apr 17;295(16):5533. doi: 10.1074/jbc.AAC120.013571. J Biol Chem. 2020. PMID: 32303645 Free PMC article. No abstract available.
Abstract
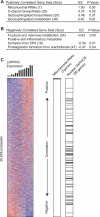
Rapid advances in genotyping and sequencing technology have dramatically accelerated the discovery of genes underlying human disease. Elucidating the function of such genes and understanding their role in pathogenesis, however, remain challenging. Here, we introduce a genomic strategy to characterize such genes functionally, and we apply it to LRPPRC, a poorly studied gene that is mutated in Leigh syndrome, French-Canadian type (LSFC). We utilize RNA interference to engineer an allelic series of cellular models in which LRPPRC has been stably silenced to different levels of knockdown efficiency. We then combine genome-wide expression profiling with gene set enrichment analysis to identify cellular responses that correlate with the loss of LRPPRC. Using this strategy, we discovered a specific role for LRPPRC in the expression of all mitochondrial DNA-encoded mRNAs, but not the rRNAs, providing mechanistic insights into the enzymatic defects observed in the disease. Our analysis shows that nuclear genes encoding mitochondrial proteins are not collectively affected by the loss of LRPPRC. We do observe altered expression of genes related to hexose metabolism, prostaglandin synthesis, and glycosphingolipid biology that may either play an adaptive role in cell survival or contribute to pathogenesis. The combination of genetic perturbation, genomic profiling, and pathway analysis represents a generic strategy for understanding disease pathogenesis.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

