Molecular epidemiology of vancomycin-resistant Enterococcus faecium: a prospective, multicenter study in South American hospitals
- PMID: 20220167
- PMCID: PMC2863891
- DOI: 10.1128/JCM.02526-09
Molecular epidemiology of vancomycin-resistant Enterococcus faecium: a prospective, multicenter study in South American hospitals
Abstract
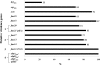
Enterococcus faecium has emerged as an important nosocomial pathogen worldwide, and this trend has been associated with the dissemination of a genetic lineage designated clonal cluster 17 (CC17). Enterococcal isolates were collected prospectively (2006 to 2008) from 32 hospitals in Colombia, Ecuador, Perú, and Venezuela and subjected to antimicrobial susceptibility testing. Genotyping was performed with all vancomycin-resistant E. faecium (VREfm) isolates by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing. All VREfm isolates were evaluated for the presence of 16 putative virulence genes (14 fms genes, the esp gene of E. faecium [espEfm], and the hyl gene of E. faecium [hylEfm]) and plasmids carrying the fms20-fms21 (pilA), hylEfm, and vanA genes. Of 723 enterococcal isolates recovered, E. faecalis was the most common (78%). Vancomycin resistance was detected in 6% of the isolates (74% of which were E. faecium). Eleven distinct PFGE types were found among the VREfm isolates, with most belonging to sequence types 412 and 18. The ebpAEfm-ebpBEfm-ebpCEfm (pilB) and fms11-fms19-fms16 clusters were detected in all VREfm isolates from the region, whereas espEfm and hylEfm were detected in 69% and 23% of the isolates, respectively. The fms20-fms21 (pilA) cluster, which encodes a putative pilus-like protein, was found on plasmids from almost all VREfm isolates and was sometimes found to coexist with hylEfm and the vanA gene cluster. The population genetics of VREfm in South America appear to resemble those of such strains in the United States in the early years of the CC17 epidemic. The overwhelming presence of plasmids encoding putative virulence factors and vanA genes suggests that E. faecium from the CC17 genogroup may disseminate in the region in the coming years.
Figures


References
-
- Arias, C. A., J. Reyes, M. Zúñiga, L. Cortes, C. Cruz, L. Rico, and D. Panesso. 2003. Multicentre surveillance of antimicrobial resistance in enterococci and staphylococci from Colombian hospitals, 2001-2002. J. Antimicrob. Chemother. 51:59-68. - PubMed
-
- Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

