Tumor-associated macrophages and survival in classic Hodgkin's lymphoma
- PMID: 20220182
- PMCID: PMC2897174
- DOI: 10.1056/NEJMoa0905680
Tumor-associated macrophages and survival in classic Hodgkin's lymphoma
Abstract
Background: Despite advances in treatments for Hodgkin's lymphoma, about 20% of patients still die from progressive disease. Current prognostic models predict the outcome of treatment with imperfect accuracy, and clinically relevant biomarkers have not been established to improve on the International Prognostic Score.
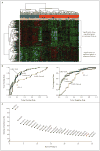
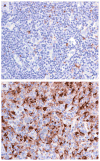
Methods: Using gene-expression profiling, we analyzed 130 frozen samples obtained from patients with classic Hodgkin's lymphoma during diagnostic lymph-node biopsy to determine which cellular signatures were correlated with treatment outcome. We confirmed our findings in an independent cohort of 166 patients, using immunohistochemical analysis.
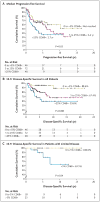
Results: Gene-expression profiling identified a gene signature of tumor-associated macrophages that was significantly associated with primary treatment failure (P=0.02). In an independent cohort of patients, we found that an increased number of CD68+ macrophages was correlated with a shortened progression-free survival (P=0.03) and with an increased likelihood of relapse after autologous hematopoietic stem-cell transplantation (P=0.008), resulting in shortened disease-specific survival (P=0.003). In multivariate analysis, this adverse prognostic factor outperformed the International Prognostic Score for disease-specific survival (P=0.003 vs. P=0.03). The absence of an elevated number of CD68+ cells in patients with limited-stage disease defined a subgroup of patients with a long-term disease-specific survival of 100% with the use of current treatment strategies.
Conclusions: An increased number of tumor-associated macrophages was strongly associated with shortened survival in patients with classic Hodgkin's lymphoma and provides a new biomarker for risk stratification.
2010 Massachusetts Medical Society
Figures
Comment in
-
Toward a personalized treatment of Hodgkin's disease.N Engl J Med. 2010 Mar 11;362(10):942-3. doi: 10.1056/NEJMe0912481. N Engl J Med. 2010. PMID: 20220189 No abstract available.
-
Macrophages in Hodgkin's lymphoma.N Engl J Med. 2010 Jun 3;362(22):2135; author reply 2136. doi: 10.1056/NEJMc1004039. N Engl J Med. 2010. PMID: 20519687 No abstract available.
-
Macrophages in Hodgkin's lymphoma.N Engl J Med. 2010 Jun 3;362(22):2135-6; author reply 2136. N Engl J Med. 2010. PMID: 20527059 No abstract available.
-
Macrophages in Hodgkin's lymphoma.N Engl J Med. 2010 Jun 3;362(22):2135; author reply 2136. N Engl J Med. 2010. PMID: 20527079 No abstract available.
References
-
- Björkholm M, Axdorph U, Grimfors G, et al. Fixed versus response-adapted MOPP/ABVD chemotherapy in Hodgkin's disease: a prospective randomized trial. Ann Oncol. 1995;6:895–9. - PubMed
-
- Canellos GP, Anderson JR, Propert KJ, et al. Chemotherapy of advanced Hodgkin's disease with MOPP, ABVD, or MOPP alternating with ABVD. N Engl J Med. 1992;327:1478–84. - PubMed
-
- Diehl V, Stein H, Hummel M, Zollinger R, Connors JM. Hodgkin's lymphoma: biology and treatment strategies for primary, refractory, and relapsed disease. Hematology Am Soc Hematol Educ Program. 2003:225–47. - PubMed
-
- Hasenclever D, Diehl V, Armitage JO, et al. A prognostic score for advanced Hodgkin's disease. N Engl J Med. 1998;339:1506–14. - PubMed
-
- Swerdlow SH, Campo E, Harris NL, et al., editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th. Lyon, France: International Agency for Research on Cancer; 2008.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
