Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models
- PMID: 20220767
- PMCID: PMC3128718
- DOI: 10.1038/jid.2010.36
Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models
Abstract
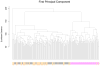
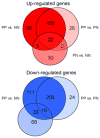
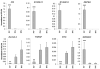
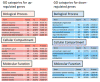
To further elucidate molecular alterations in psoriasis, we performed a gene expression study of 58 paired lesional and uninvolved psoriatic and 64 control skin samples. Comparison of involved psoriatic (PP) and normal (NN) skin identified 1,326 differentially regulated transcripts encoding 918 unique genes (549 up- and 369 downregulated), of which over 600 are to our knowledge previously unreported, including S100A7A, THRSP, and ELOVL3. Strongly upregulated genes included SERPINB4, PI3, DEFB4, and several S100-family members. Strongly downregulated genes included Wnt-inhibitory factor-1 (WIF1), beta-cellulin (BTC), and CCL27. Enriched gene ontology categories included immune response, defense response, and keratinocyte differentiation. Biological processes regulating fatty acid and lipid metabolism were enriched in the down-regulated gene set. Comparison of the psoriatic transcriptome to the transcriptomes of cytokine-stimulated cultured keratinocytes (IL-17, IL-22, IL-1alpha, IFN-gamma, TNF-alpha, and OSM) showed surprisingly little overlap, with the cytokine-stimulated keratinocyte expression representing only 2.5, 0.7, 1.5, 5.6, 5.0, and 1.9% of the lesional psoriatic dysregulated transcriptome, respectively. This comprehensive analysis of differentially regulated transcripts in psoriasis provides additional insight into the pathogenic mechanisms involved and emphasizes the need for more complex yet tractable experimental models of psoriasis.
Conflict of interest statement
Figures

References
-
- Akey JM, Biswas S, Leek JT, Storey JD. On the design and analysis of gene expression studies in human populations. Nat Genet. 2007;39:807–8. author reply 8-9. - PubMed
-
- Alkemade JA, Molhuizen HO, Ponec M, Kempenaar JA, Zeeuwen PL, de Jongh GJ, et al. SKALP/elafin is an inducible proteinase inhibitor in human epidermal keratinocytes. J Cell Sci. 1994;107(Pt 8):2335–42. - PubMed
-
- Bando M, Hiroshima Y, Kataoka M, Shinohara Y, Herzberg MC, Ross KF, et al. Interleukin-1alpha regulates antimicrobial peptide expression in human keratinocytes. Immunol Cell Biol. 2007;85:532–7. - PubMed
-
- Banno T, Adachi M, Mukkamala L, Blumenberg M. Unique keratinocyte-specific effects of interferon-gamma that protect skin from viruses, identified using transcriptional profiling. Antivir Ther. 2003;8:541–54. - PubMed
-
- Banno T, Gazel A, Blumenberg M. Effects of tumor necrosis factor-alpha (TNF alpha) in epidermal keratinocytes revealed using global transcriptional profiling. J Biol Chem. 2004;279:32633–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

