Quantitative lymphatic vessel trait analysis suggests Vcam1 as candidate modifier gene of inflammatory bowel disease
- PMID: 20220769
- PMCID: PMC2865135
- DOI: 10.1038/gene.2010.4
Quantitative lymphatic vessel trait analysis suggests Vcam1 as candidate modifier gene of inflammatory bowel disease
Abstract
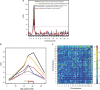
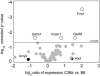
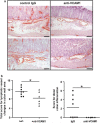
Inflammatory bowel disease (IBD) is a chronic debilitating disease resulting from a complex interaction of multiple genetic factors with the environment. To identify modifier genes of IBD, we used an F2 intercross of IBD-resistant C57BL/6J-Il10(-/-) mice and IBD-susceptible C3H/HeJBir-Il10(-/-) (C3Bir-Il10(-/-)) mice. We found a prominent involvement of lymphatic vessels in IBD and applied a scoring system to quantify lymphatic vascular changes. Quantitative trait locus (QTL) analyses revealed a large-effect QTL on chromosome 3, mapping to an interval of 43.6 Mbp. This candidate interval was narrowed by fine mapping to 22 Mbp, and candidate genes were analyzed by a systems genetics approach that included quantitative gene expression profiling, search for functional polymorphisms, and haplotype block analysis. We identified vascular adhesion molecule 1 (Vcam1) as a candidate modifier gene in the interleukin 10-deficient mouse model of IBD. Importantly, VCAM1 protein levels were increased in susceptible C3H/HeJ mice, compared with C57BL/6J mice; systemic blockade of VCAM1 in C3Bir-Il10(-/-) mice reduced their inflammatory lymphatic vessel changes. These results indicate that genetically determined expression differences of VCAM1 are associated with susceptibility to colon inflammation, which is accompanied by extensive lymphatic vessel changes. VCAM1 is, therefore, a promising therapeutic target for IBD.
Figures




Similar articles
-
Cd14, Gbp1, and Pla2g2a: three major candidate genes for experimental IBD identified by combining QTL and microarray analyses.Physiol Genomics. 2006 May 16;25(3):426-34. doi: 10.1152/physiolgenomics.00022.2005. Physiol Genomics. 2006. PMID: 16705022
-
Analysis of Cd14 as a genetic modifier of experimental inflammatory bowel disease (IBD) in mice.Inflamm Bowel Dis. 2009 Dec;15(12):1824-36. doi: 10.1002/ibd.21030. Epub 2009 Jul 27. Inflamm Bowel Dis. 2009. PMID: 19637338
-
Cdcs1 a major colitis susceptibility locus in mice; subcongenic analysis reveals genetic complexity.Inflamm Bowel Dis. 2010 May;16(5):765-75. doi: 10.1002/ibd.21146. Inflamm Bowel Dis. 2010. PMID: 19856416 Free PMC article.
-
Genetic and environmental context determines the course of colitis developing in IL-10-deficient mice.Inflamm Bowel Dis. 2002 Sep;8(5):347-55. doi: 10.1097/00054725-200209000-00006. Inflamm Bowel Dis. 2002. PMID: 12479650 Review.
-
Mapping colitis susceptibility in mouse models: distal chromosome 3 contains major loci related to Cdcs1.Physiol Genomics. 2013 Oct 16;45(20):925-30. doi: 10.1152/physiolgenomics.00084.2013. Epub 2013 Sep 10. Physiol Genomics. 2013. PMID: 24022218 Review.
Cited by
-
Quantitative Trait Locus and Integrative Genomics Revealed Candidate Modifier Genes for Ectopic Mineralization in Mouse Models of Pseudoxanthoma Elasticum.J Invest Dermatol. 2019 Dec;139(12):2447-2457.e7. doi: 10.1016/j.jid.2019.04.023. Epub 2019 Jun 15. J Invest Dermatol. 2019. PMID: 31207231 Free PMC article.
-
Blockade of VEGF receptor-3 aggravates inflammatory bowel disease and lymphatic vessel enlargement.Inflamm Bowel Dis. 2013 Aug;19(9):1983-9. doi: 10.1097/MIB.0b013e31829292f7. Inflamm Bowel Dis. 2013. PMID: 23835443 Free PMC article.
-
Obstructive Lymphangitis Precedes Colitis in Murine Norovirus-Infected Stat1-Deficient Mice.Am J Pathol. 2018 Jul;188(7):1536-1554. doi: 10.1016/j.ajpath.2018.03.019. Epub 2018 May 18. Am J Pathol. 2018. PMID: 29753791 Free PMC article.
-
Thymus cell antigen 1 (Thy1, CD90) is expressed by lymphatic vessels and mediates cell adhesion to lymphatic endothelium.Exp Cell Res. 2010 Oct 15;316(17):2982-92. doi: 10.1016/j.yexcr.2010.06.013. Epub 2010 Jun 23. Exp Cell Res. 2010. PMID: 20599951 Free PMC article.
-
A Multihit Model: Colitis Lessons from the Interleukin-10-deficient Mouse.Inflamm Bowel Dis. 2015 Aug;21(8):1967-75. doi: 10.1097/MIB.0000000000000468. Inflamm Bowel Dis. 2015. PMID: 26164667 Free PMC article. Review.
References
-
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. - PubMed
-
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. - PubMed
-
- Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

