Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm
- PMID: 20221255
- PMCID: PMC2832674
- DOI: 10.1371/journal.pcbi.1000694
Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm
Abstract
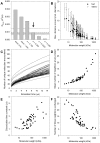
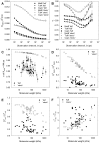
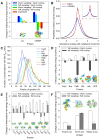
A longstanding question in molecular biology is the extent to which the behavior of macromolecules observed in vitro accurately reflects their behavior in vivo. A number of sophisticated experimental techniques now allow the behavior of individual types of macromolecule to be studied directly in vivo; none, however, allow a wide range of molecule types to be observed simultaneously. In order to tackle this issue we have adopted a computational perspective, and, having selected the model prokaryote Escherichia coli as a test system, have assembled an atomically detailed model of its cytoplasmic environment that includes 50 of the most abundant types of macromolecules at experimentally measured concentrations. Brownian dynamics (BD) simulations of the cytoplasm model have been calibrated to reproduce the translational diffusion coefficients of Green Fluorescent Protein (GFP) observed in vivo, and "snapshots" of the simulation trajectories have been used to compute the cytoplasm's effects on the thermodynamics of protein folding, association and aggregation events. The simulation model successfully describes the relative thermodynamic stabilities of proteins measured in E. coli, and shows that effects additional to the commonly cited "crowding" effect must be included in attempts to understand macromolecular behavior in vivo.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Making Soup: Preparing and Validating Models of the Bacterial Cytoplasm for Molecular Simulation.J Chem Inf Model. 2020 Jan 27;60(1):322-331. doi: 10.1021/acs.jcim.9b00971. Epub 2019 Dec 19. J Chem Inf Model. 2020. PMID: 31816234
-
Atomically detailed simulations of concentrated protein solutions: the effects of salt, pH, point mutations, and protein concentration in simulations of 1000-molecule systems.J Am Chem Soc. 2006 Sep 20;128(37):12098-110. doi: 10.1021/ja0614058. J Am Chem Soc. 2006. PMID: 16967959
-
An analytical correlated random walk model and its application to understand subdiffusion in crowded environment.J Chem Phys. 2015 Sep 21;143(11):114104. doi: 10.1063/1.4930275. J Chem Phys. 2015. PMID: 26395684
-
Recent Progress in Modeling and Simulation of Biomolecular Crowding and Condensation Inside Cells.J Chem Inf Model. 2024 Dec 23;64(24):9063-9081. doi: 10.1021/acs.jcim.4c01520. Epub 2024 Dec 11. J Chem Inf Model. 2024. PMID: 39660892 Free PMC article. Review.
-
Biochemical effects of molecular crowding.Biochemistry (Mosc). 2004 Nov;69(11):1239-51. doi: 10.1007/s10541-005-0070-y. Biochemistry (Mosc). 2004. PMID: 15627378 Review.
Cited by
-
Perspective: On the importance of hydrodynamic interactions in the subcellular dynamics of macromolecules.J Chem Phys. 2016 Sep 14;145(10):100901. doi: 10.1063/1.4962258. J Chem Phys. 2016. PMID: 27634243 Free PMC article.
-
Enzyme activity in the crowded milieu.PLoS One. 2012;7(6):e39418. doi: 10.1371/journal.pone.0039418. Epub 2012 Jun 26. PLoS One. 2012. PMID: 22761790 Free PMC article.
-
Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs.RNA. 2016 Jun;22(6):896-904. doi: 10.1261/rna.055632.115. Epub 2016 Apr 18. RNA. 2016. PMID: 27090284 Free PMC article.
-
Multiscale simulation of molecular processes in cellular environments.Philos Trans A Math Phys Eng Sci. 2016 Nov 13;374(2080):20160225. doi: 10.1098/rsta.2016.0225. Philos Trans A Math Phys Eng Sci. 2016. PMID: 27698046 Free PMC article. Review.
-
Analysis of Fluorescent Proteins for Observing Single Gene Locus in a Live and Fixed Escherichia coli Cell.J Phys Chem B. 2024 Jul 18;128(28):6730-6741. doi: 10.1021/acs.jpcb.4c01816. Epub 2024 Jul 5. J Phys Chem B. 2024. PMID: 38968413 Free PMC article.
References
-
- Dix JA, Verkman AS. Crowding effects on diffusion in solutions and cells. Annu Rev Biophys. 2008;37:247–263. - PubMed
-
- Zimmerman SB, Minton AP. Macromolecular crowding – biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. - PubMed
-
- Ghaemmaghami S, Oas TG. Quantitative protein stability measurement in vivo. Nature Struct Biol. 2001;8:879–882. - PubMed
-
- Reckel S, Hänsel R, Löhr F, Dötsch V. In-cell NMR spectroscopy. Prog Nuc Mag Res Spec. 2007;51:91–101.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

