Mathematical modelling of cell-fate decision in response to death receptor engagement
- PMID: 20221256
- PMCID: PMC2832675
- DOI: 10.1371/journal.pcbi.1000702
Mathematical modelling of cell-fate decision in response to death receptor engagement
Abstract
Cytokines such as TNF and FASL can trigger death or survival depending on cell lines and cellular conditions. The mechanistic details of how a cell chooses among these cell fates are still unclear. The understanding of these processes is important since they are altered in many diseases, including cancer and AIDS. Using a discrete modelling formalism, we present a mathematical model of cell fate decision recapitulating and integrating the most consistent facts extracted from the literature. This model provides a generic high-level view of the interplays between NFkappaB pro-survival pathway, RIP1-dependent necrosis, and the apoptosis pathway in response to death receptor-mediated signals. Wild type simulations demonstrate robust segregation of cellular responses to receptor engagement. Model simulations recapitulate documented phenotypes of protein knockdowns and enable the prediction of the effects of novel knockdowns. In silico experiments simulate the outcomes following ligand removal at different stages, and suggest experimental approaches to further validate and specialise the model for particular cell types. We also propose a reduced conceptual model implementing the logic of the decision process. This analysis gives specific predictions regarding cross-talks between the three pathways, as well as the transient role of RIP1 protein in necrosis, and confirms the phenotypes of novel perturbations. Our wild type and mutant simulations provide novel insights to restore apoptosis in defective cells. The model analysis expands our understanding of how cell fate decision is made. Moreover, our current model can be used to assess contradictory or controversial data from the literature. Ultimately, it constitutes a valuable reasoning tool to delineate novel experiments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
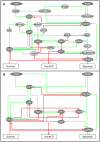




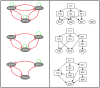
References
-
- Rehm M, Huber HJ, Hellwig CT, Anguissola S, Dussmann H, et al. Dynamics of outer mitochondrial membrane permeabilization during apoptosis. Cell Death Differ. 2009;16:613–623. - PubMed
-
- Eissing T, Conzelmann H, Gilles ED, Allgower F, Bullinger E, et al. Bistability analyses of a caspase activation model for receptor-induced apoptosis. J Biol Chem. 2004;279:36892–36897. - PubMed
-
- Fussenegger M, Bailey JE, Varner J. A mathematical model of caspase function in apoptosis. Nat Biotechnol. 2000;18:768–774. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

