Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish
- PMID: 20221429
- PMCID: PMC2832694
- DOI: 10.1371/journal.pone.0009478
Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish
Abstract
Background: Jawed vertebrates generate their immune-receptor repertoire by a recombinatorial mechanism that has the potential to produce harmful autoreactive lymphocytes. In mammals, peripheral tolerance to self-antigens is enforced by Foxp3(+) regulatory T cells. Recombinatorial mechanisms also operate in teleosts, but active immunoregulation is thought to be a late incorporation to the vertebrate lineage.
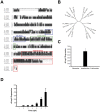
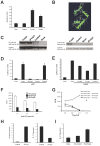
Methods/principal findings: Here we report the characterization of adaptive autoimmunity and Foxp3-based immunoregulation in the zebrafish. We found that zebrafish immunization with an homogenate of zebrafish central nervous system (zCNS) triggered CNS inflammation and specific antibodies. We cloned the zebrafish ortholog for mammalian Foxp3 (zFoxp3) which induced a regulatory phenotype on mouse T cells and controlled IL-17 production in zebrafish embryos.
Conclusions/significance: Our findings demonstrate the acquisition of active mechanisms of self-tolerance early in vertebrate evolution, suggesting that active regulatory mechanisms accompany the development of the molecular potential for adaptive autoimmunity. Moreover, they identify the zebrafish as a tool to study the molecular pathways controlling adaptive immunity.
Conflict of interest statement
Figures



References
-
- Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. - PubMed
-
- Honjo T, Nagaoka H, Shinkura R, Muramatsu M. AID to overcome the limitations of genomic information. Nat Immunol. 2005;6:655–661. - PubMed
-
- Bouaziz JD, Yanaba K, Tedder TF. Regulatory B cells as inhibitors of immune responses and inflammation. Immunol Rev. 2008;224:201–214. - PubMed
-
- Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. - PubMed
-
- Roncarolo MG, Battaglia M. Regulatory T-cell immunotherapy for tolerance to self antigens and alloantigens in humans. Nat Rev Immunol. 2007;7:585–598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

