Array-based FMR1 sequencing and deletion analysis in patients with a fragile X syndrome-like phenotype
- PMID: 20221430
- PMCID: PMC2832695
- DOI: 10.1371/journal.pone.0009476
Array-based FMR1 sequencing and deletion analysis in patients with a fragile X syndrome-like phenotype
Abstract
Background: Fragile X syndrome (FXS) is caused by loss of function mutations in the FMR1 gene. Trinucleotide CGG-repeat expansions, resulting in FMR1 gene silencing, are the most common mutations observed at this locus. Even though the repeat expansion mutation is a functional null mutation, few conventional mutations have been identified at this locus, largely due to the clinical laboratory focus on the repeat tract.
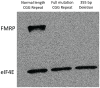
Methodology/principal findings: To more thoroughly evaluate the frequency of conventional mutations in FXS-like patients, we used an array-based method to sequence FMR1 in 51 unrelated males exhibiting several features characteristic of FXS but with normal CGG-repeat tracts of FMR1. One patient was identified with a deletion in FMR1, but none of the patients were found to have other conventional mutations.
Conclusions/significance: These data suggest that missense mutations in FMR1 are not a common cause of the FXS phenotype in patients who have normal-length CGG-repeat tracts. However, screening for small deletions of FMR1 may be of clinically utility.
Conflict of interest statement
Figures
Similar articles
-
Point mutation frequency in the FMR1 gene as revealed by fragile X syndrome screening.Mol Cell Probes. 2014 Oct-Dec;28(5-6):279-83. doi: 10.1016/j.mcp.2014.08.003. Epub 2014 Aug 27. Mol Cell Probes. 2014. PMID: 25171808
-
CGG-repeat dynamics and FMR1 gene silencing in fragile X syndrome stem cells and stem cell-derived neurons.Mol Autism. 2016 Oct 6;7:42. doi: 10.1186/s13229-016-0105-9. eCollection 2016. Mol Autism. 2016. PMID: 27713816 Free PMC article.
-
FMR1 CGG repeat lengths mediate different regulation of reporter gene expression in comparative transient and locus specific integration assays.Gene. 2011 Oct 15;486(1-2):15-22. doi: 10.1016/j.gene.2011.06.034. Epub 2011 Jul 13. Gene. 2011. PMID: 21767618
-
Fragile X syndrome and deletions in FMR1: new case and review of the literature.Am J Med Genet. 1997 Nov 12;72(4):430-4. Am J Med Genet. 1997. PMID: 9375726 Review.
-
Spontaneous rescue of a FMR1 repeat expansion and review of deletions in the FMR1 non-coding region.Eur J Med Genet. 2021 Aug;64(8):104244. doi: 10.1016/j.ejmg.2021.104244. Epub 2021 May 20. Eur J Med Genet. 2021. PMID: 34022415 Review.
Cited by
-
The fragile x mental retardation syndrome 20 years after the FMR1 gene discovery: an expanding universe of knowledge.Clin Biochem Rev. 2011 Aug;32(3):135-62. Clin Biochem Rev. 2011. PMID: 21912443 Free PMC article.
-
Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males.Am J Med Genet A. 2010 Oct;152A(10):2512-20. doi: 10.1002/ajmg.a.33626. Am J Med Genet A. 2010. PMID: 20799337 Free PMC article.
-
Beyond Trinucleotide Repeat Expansion in Fragile X Syndrome: Rare Coding and Noncoding Variants in FMR1 and Associated Phenotypes.Genes (Basel). 2021 Oct 22;12(11):1669. doi: 10.3390/genes12111669. Genes (Basel). 2021. PMID: 34828275 Free PMC article.
-
EMQN best practice guidelines for the molecular genetic testing and reporting of fragile X syndrome and other fragile X-associated disorders.Eur J Hum Genet. 2015 Apr;23(4):417-25. doi: 10.1038/ejhg.2014.185. Epub 2014 Sep 17. Eur J Hum Genet. 2015. PMID: 25227148 Free PMC article.
-
Molecular Correlates and Recent Advancements in the Diagnosis and Screening of FMR1-Related Disorders.Genes (Basel). 2016 Oct 14;7(10):87. doi: 10.3390/genes7100087. Genes (Basel). 2016. PMID: 27754417 Free PMC article. Review.
References
-
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. - PubMed
-
- Gedeon AK, Baker E, Robinson H, Partington MW, Gross B, et al. Fragile X syndrome without CCG amplification has an FMR1 deletion. Nat Genet. 1992;1:341–344. - PubMed
-
- Tarleton J, Richie R, Schwartz C, Rao K, Aylsworth AS, et al. An extensive de novo deletion removing FMR1 in a patient with mental retardation and the fragile X syndrome phenotype. Hum Mol Genet. 1993;2:1973–1974. - PubMed
-
- Lugenbeel KA, Peier AM, Carson NL, Chudley AE, Nelson DL. Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat Genet. 1995;10:483–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

