Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene
- PMID: 20221433
- PMCID: PMC2832698
- DOI: 10.1371/journal.pone.0009545
Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene
Abstract
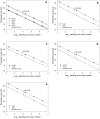
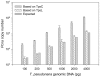
Quantitative real-time PCR (qPCR) has become a gold standard for the quantification of nucleic acids and microorganism abundances, in which plasmid DNA carrying the target genes are most commonly used as the standard. A recent study showed that supercoiled circular confirmation of DNA appeared to suppress PCR amplification. However, to what extent to which different structural types of DNA (circular versus linear) used as the standard may affect the quantification accuracy has not been evaluated. In this study, we quantitatively compared qPCR accuracies based on circular plasmid (mostly in supercoiled form) and linear DNA standards (linearized plasmid DNA or PCR amplicons), using proliferating cell nuclear gene (pcna), the ubiquitous eukaryotic gene, in five marine microalgae as a model gene. We observed that PCR using circular plasmids as template gave 2.65-4.38 more of the threshold cycle number than did equimolar linear standards. While the documented genome sequence of the diatom Thalassiosira pseudonana shows a single copy of pcna, qPCR using the circular plasmid as standard yielded an estimate of 7.77 copies of pcna per genome whereas that using the linear standard gave 1.02 copies per genome. We conclude that circular plasmid DNA is unsuitable as a standard, and linear DNA should be used instead, in absolute qPCR. The serious overestimation by the circular plasmid standard is likely due to the undetected lower efficiency of its amplification in the early stage of PCR when the supercoiled plasmid is the dominant template.
Conflict of interest statement
Figures
Similar articles
-
Similar gene estimates from circular and linear standards in quantitative PCR analyses using the prokaryotic 16S rRNA gene as a model.PLoS One. 2012;7(12):e51931. doi: 10.1371/journal.pone.0051931. Epub 2012 Dec 19. PLoS One. 2012. PMID: 23284822 Free PMC article.
-
Enhancement of PCR Detection Limit by Single-Tube Restriction Endonuclease-PCR (RE-PCR).Mol Diagn Ther. 2016 Jun;20(3):297-305. doi: 10.1007/s40291-016-0195-2. Mol Diagn Ther. 2016. PMID: 26993322
-
Quantification bias caused by plasmid DNA conformation in quantitative real-time PCR assay.PLoS One. 2011;6(12):e29101. doi: 10.1371/journal.pone.0029101. Epub 2011 Dec 14. PLoS One. 2011. PMID: 22194997 Free PMC article.
-
Molecular detection, quantification, and diversity evaluation of microalgae.Mar Biotechnol (NY). 2012 Apr;14(2):129-42. doi: 10.1007/s10126-011-9427-y. Epub 2011 Dec 28. Mar Biotechnol (NY). 2012. PMID: 22200918 Review.
-
[Quantitative PCR in the diagnosis of Leishmania].Parassitologia. 2004 Jun;46(1-2):163-7. Parassitologia. 2004. PMID: 15305709 Review. Italian.
Cited by
-
Induction of lcc2 expression and activity by Agaricus bisporus provides defence against Trichoderma aggressivum toxic extracts.Microb Biotechnol. 2015 Nov;8(6):918-29. doi: 10.1111/1751-7915.12277. Epub 2015 Mar 30. Microb Biotechnol. 2015. PMID: 25824278 Free PMC article.
-
Newcastle disease virus genotype VII gene expression in experimentally infected birds.Sci Rep. 2022 Mar 28;12(1):5249. doi: 10.1038/s41598-022-09257-y. Sci Rep. 2022. PMID: 35347193 Free PMC article.
-
Accumulation and Competition Amongst Deformed Wing Virus Genotypes in Naïve Australian Honeybees Provides Insight Into the Increasing Global Prevalence of Genotype B.Front Microbiol. 2020 Apr 9;11:620. doi: 10.3389/fmicb.2020.00620. eCollection 2020. Front Microbiol. 2020. PMID: 32328051 Free PMC article.
-
Tandem repeats, high copy number and remarkable diel expression rhythm of form II RuBisCO in Prorocentrum donghaiense (Dinophyceae).PLoS One. 2013 Aug 19;8(8):e71232. doi: 10.1371/journal.pone.0071232. eCollection 2013. PLoS One. 2013. PMID: 23976999 Free PMC article.
-
Accurate Quantification of AAV Vector Genomes by Quantitative PCR.Genes (Basel). 2021 Apr 19;12(4):601. doi: 10.3390/genes12040601. Genes (Basel). 2021. PMID: 33921790 Free PMC article.
References
-
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. - PubMed
-
- Freeman WM, Walker SJ, Vrana KE. Quantitative RT-PCR: Pitfalls and potential. Biotechniques. 1999;26:112–125. - PubMed
-
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. - PubMed
-
- Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med. 2002;8:257–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

