Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species
- PMID: 20221447
- PMCID: PMC2833190
- DOI: 10.1371/journal.pone.0009556
Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species
Abstract
Background: The genus Cronobacter (formerly called Enterobacter sakazakii) is composed of five species; C. sakazakii, C. malonaticus, C. turicensis, C. muytjensii, and C. dublinensis. The genus includes opportunistic human pathogens, and the first three species have been associated with neonatal infections. The most severe diseases are caused in neonates and include fatal necrotizing enterocolitis and meningitis. The genetic basis of the diversity within the genus is unknown, and few virulence traits have been identified.
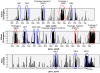
Methodology/principal findings: We report here the first sequence of a member of this genus, C. sakazakii strain BAA-894. The genome of Cronobacter sakazakii strain BAA-894 comprises a 4.4 Mb chromosome (57% GC content) and two plasmids; 31 kb (51% GC) and 131 kb (56% GC). The genome was used to construct a 387,000 probe oligonucleotide tiling DNA microarray covering the whole genome. Comparative genomic hybridization (CGH) was undertaken on five other C. sakazakii strains, and representatives of the four other Cronobacter species. Among 4,382 annotated genes inspected in this study, about 55% of genes were common to all C. sakazakii strains and 43% were common to all Cronobacter strains, with 10-17% absence of genes.
Conclusions/significance: CGH highlighted 15 clusters of genes in C. sakazakii BAA-894 that were divergent or absent in more than half of the tested strains; six of these are of probable prophage origin. Putative virulence factors were identified in these prophage and in other variable regions. A number of genes unique to Cronobacter species associated with neonatal infections (C. sakazakii, C. malonaticus and C. turicensis) were identified. These included a copper and silver resistance system known to be linked to invasion of the blood-brain barrier by neonatal meningitic strains of Escherichia coli. In addition, genes encoding for multidrug efflux pumps and adhesins were identified that were unique to C. sakazakii strains from outbreaks in neonatal intensive care units.
Conflict of interest statement
Figures


Similar articles
-
Microarray-based comparative genomic indexing of the Cronobacter genus (Enterobacter sakazakii).Int J Food Microbiol. 2009 Dec 31;136(2):159-64. doi: 10.1016/j.ijfoodmicro.2009.07.008. Epub 2009 Jul 13. Int J Food Microbiol. 2009. PMID: 19664834
-
Comparative analysis of genome sequences covering the seven cronobacter species.PLoS One. 2012;7(11):e49455. doi: 10.1371/journal.pone.0049455. Epub 2012 Nov 16. PLoS One. 2012. PMID: 23166675 Free PMC article.
-
Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov.Int J Syst Evol Microbiol. 2008 Jun;58(Pt 6):1442-7. doi: 10.1099/ijs.0.65577-0. Int J Syst Evol Microbiol. 2008. PMID: 18523192
-
Cronobacter spp.--opportunistic food-borne pathogens. A review of their virulence and environmental-adaptive traits.J Med Microbiol. 2014 Aug;63(Pt 8):1023-1037. doi: 10.1099/jmm.0.073742-0. Epub 2014 May 30. J Med Microbiol. 2014. PMID: 24878566 Review.
-
Insights into virulence factors determining the pathogenicity of Cronobacter sakazakii.Virulence. 2015;6(5):433-40. doi: 10.1080/21505594.2015.1036217. Epub 2015 May 7. Virulence. 2015. PMID: 25950947 Free PMC article. Review.
Cited by
-
Cronobacter Species Contamination of Powdered Infant Formula and the Implications for Neonatal Health.Front Pediatr. 2015 Jul 2;3:56. doi: 10.3389/fped.2015.00056. eCollection 2015. Front Pediatr. 2015. PMID: 26191519 Free PMC article. Review.
-
Reconstruction of the carotenoid biosynthetic pathway of Cronobacter sakazakii BAA894 in Escherichia coli.PLoS One. 2014 Jan 23;9(1):e86739. doi: 10.1371/journal.pone.0086739. eCollection 2014. PLoS One. 2014. PMID: 24466219 Free PMC article.
-
Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis.BMC Genomics. 2014 Dec 16;15(1):1121. doi: 10.1186/1471-2164-15-1121. BMC Genomics. 2014. PMID: 25515150 Free PMC article.
-
Comparative Genotypic and Phenotypic Analysis of Cronobacter Species Cultured from Four Powdered Infant Formula Production Facilities: Indication of Pathoadaptation along the Food Chain.Appl Environ Microbiol. 2015 Jul;81(13):4388-402. doi: 10.1128/AEM.00359-15. Epub 2015 Apr 24. Appl Environ Microbiol. 2015. PMID: 25911470 Free PMC article.
-
Multiplex PCR assay targeting a diguanylate cyclase-encoding gene, cgcA, to differentiate species within the genus Cronobacter.Appl Environ Microbiol. 2013 Jan;79(2):734-7. doi: 10.1128/AEM.02898-12. Epub 2012 Nov 9. Appl Environ Microbiol. 2013. PMID: 23144142 Free PMC article.
References
-
- Friedemann M. Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int J Food Microbiol. 2007;116:1–10. - PubMed
-
- Kandhai MC, Reij MW, Gorris LG, Guillaume-Gentil O, van Schothorst M. Occurrence of Enterobacter sakazakii in food production environments and households. Lancet. 2004;363(9402):39–40. - PubMed
-
- Gurtler JB, Kornacki JL, Beuchat LR. Enterobacter sakazakii: a coliform of increased concern to infant health. Int J Food Microbiol. 2005;104:1–34. - PubMed
-
- Lai KK. Enterobacter sakazakii infections among neonates, infants, children, and adults. Medicine. 2001;80:113–122. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

