Genome-wide functional analysis of human 5' untranslated region introns
- PMID: 20222956
- PMCID: PMC2864569
- DOI: 10.1186/gb-2010-11-3-r29
Genome-wide functional analysis of human 5' untranslated region introns
Abstract
Background: Approximately 35% of human genes contain introns within the 5' untranslated region (UTR). Introns in 5'UTRs differ from those in coding regions and 3'UTRs with respect to nucleotide composition, length distribution and density. Despite their presumed impact on gene regulation, the evolution and possible functions of 5'UTR introns remain largely unexplored.
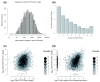
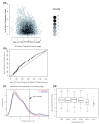
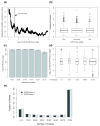
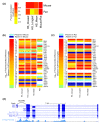
Results: We performed a genome-scale computational analysis of 5'UTR introns in humans. We discovered that the most highly expressed genes tended to have short 5'UTR introns rather than having long 5'UTR introns or lacking 5'UTR introns entirely. Although we found no correlation in 5'UTR intron presence or length with variance in expression across tissues, which might have indicated a broad role in expression-regulation, we observed an uneven distribution of 5'UTR introns amongst genes in specific functional categories. In particular, genes with regulatory roles were surprisingly enriched in having 5'UTR introns. Finally, we analyzed the evolution of 5'UTR introns in non-receptor protein tyrosine kinases (NRTK), and identified a conserved DNA motif enriched within the 5'UTR introns of human NRTKs.
Conclusions: Our results suggest that human 5'UTR introns enhance the expression of some genes in a length-dependent manner. While many 5'UTR introns are likely to be evolving neutrally, their relationship with gene expression and overrepresentation among regulatory genes, taken together, suggest that complex evolutionary forces are acting on this distinct class of introns.
Figures

Similar articles
-
Effect of 5'UTR introns on gene expression in Arabidopsis thaliana.BMC Genomics. 2006 May 19;7:120. doi: 10.1186/1471-2164-7-120. BMC Genomics. 2006. PMID: 16712733 Free PMC article.
-
Spliceosomal introns in the 5' untranslated region of plant BTL RING-H2 ubiquitin ligases are evolutionary conserved and required for gene expression.BMC Plant Biol. 2013 Nov 14;13:179. doi: 10.1186/1471-2229-13-179. BMC Plant Biol. 2013. PMID: 24228887 Free PMC article.
-
Size-dependent enhancement of gene expression by Plasmodium 5'UTR introns.Parasit Vectors. 2024 May 27;17(1):238. doi: 10.1186/s13071-024-06319-0. Parasit Vectors. 2024. PMID: 38802937 Free PMC article.
-
The Functional Meaning of 5'UTR in Protein-Coding Genes.Int J Mol Sci. 2023 Feb 3;24(3):2976. doi: 10.3390/ijms24032976. Int J Mol Sci. 2023. PMID: 36769304 Free PMC article. Review.
-
Recent insights and novel bioinformatics tools to understand the role of microRNAs binding to 5' untranslated region.Int J Mol Sci. 2012 Dec 27;14(1):480-95. doi: 10.3390/ijms14010480. Int J Mol Sci. 2012. PMID: 23271365 Free PMC article. Review.
Cited by
-
The genomic features that affect the lengths of 5' untranslated regions in multicellular eukaryotes.BMC Bioinformatics. 2011 Oct 5;12 Suppl 9(Suppl 9):S3. doi: 10.1186/1471-2105-12-S9-S3. BMC Bioinformatics. 2011. PMID: 22152105 Free PMC article.
-
Flipons and small RNAs accentuate the asymmetries of pervasive transcription by the reset and sequence-specific microcoding of promoter conformation.J Biol Chem. 2023 Sep;299(9):105140. doi: 10.1016/j.jbc.2023.105140. Epub 2023 Aug 5. J Biol Chem. 2023. PMID: 37544644 Free PMC article. Review.
-
5' Untranslated Region Elements Show High Abundance and Great Variability in Homologous ABCA Subfamily Genes.Int J Mol Sci. 2020 Nov 23;21(22):8878. doi: 10.3390/ijms21228878. Int J Mol Sci. 2020. PMID: 33238634 Free PMC article.
-
Efficient evidence-based genome annotation with EviAnn.bioRxiv [Preprint]. 2025 May 12:2025.05.07.652745. doi: 10.1101/2025.05.07.652745. bioRxiv. 2025. PMID: 40463080 Free PMC article. Preprint.
-
UPF1 regulates myeloid cell functions and S100A9 expression by the hnRNP E2/miRNA-328 balance.Sci Rep. 2016 Aug 30;6:31995. doi: 10.1038/srep31995. Sci Rep. 2016. PMID: 27573788 Free PMC article.
References
-
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

