Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination
- PMID: 20223197
- PMCID: PMC2843397
- DOI: 10.1016/j.neuron.2010.01.027
Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination
Abstract
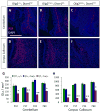
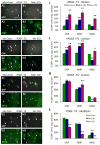
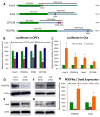
To investigate the role of microRNAs in regulating oligodendrocyte (OL) differentiation and myelination, we utilized transgenic mice in which microRNA processing was disrupted in OL precursor cells (OPCs) and OLs by targeted deletion of Dicer1. We found that inhibition of OPC-OL miRNA processing disrupts normal CNS myelination and that OPCs lacking mature miRNAs fail to differentiate normally in vitro. We identified three miRNAs (miR-219, miR-138, and miR-338) that are induced 10-100x during OL differentiation; the most strongly induced of these, miR-219, is necessary and sufficient to promote OL differentiation, and partially rescues OL differentiation defects caused by total miRNA loss. miR-219 directly represses the expression of PDGFRalpha, Sox6, FoxJ3, and ZFP238 proteins, all of which normally help to promote OPC proliferation. Together, these findings show that miR-219 plays a critical role in coupling differentiation to proliferation arrest in the OL lineage, enabling the rapid transition from proliferating OPCs to myelinating OLs.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures




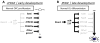
Comment in
-
Oligodendrocytes and the "micro brake" of progenitor cell proliferation.Neuron. 2010 Mar 11;65(5):577-9. doi: 10.1016/j.neuron.2010.02.026. Neuron. 2010. PMID: 20223193
-
Small RNAs: unwrapping glial differentiation.Nat Rev Neurosci. 2010 May;11(5):298. doi: 10.1038/nrn2838. Nat Rev Neurosci. 2010. PMID: 20419863 No abstract available.
References
-
- Aoki K, Meng G, Suzuki K, Takashi T, Kameoka Y, Nakahara K, Ishida R, Kasai M. RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J Biol Chem. 1998;273:26698–26704. - PubMed
-
- Assanah MC, Bruce JN, Suzuki SO, Chen A, Goldman JE, Canoll P. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia 2009 - PubMed
-
- Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994a;120:1097–1108. - PubMed
-
- Barres BA, Raff MC. Control of oligodendrocyte number in the developing rat optic nerve. Neuron. 1994;12:935–942. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

