Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail
- PMID: 20223770
- PMCID: PMC2910041
- DOI: 10.1093/nar/gkq147
Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail
Abstract
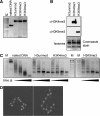
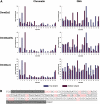
Using peptide arrays and binding to native histone proteins, we show that the ADD domain of Dnmt3a specifically interacts with the H3 histone 1-19 tail. Binding is disrupted by di- and trimethylation of K4, phosphorylation of T3, S10 or T11 and acetylation of K4. We did not observe binding to the H4 1-19 tail. The ADD domain of Dnmt3b shows the same binding specificity, suggesting that the distinct biological functions of both enzymes are not related to their ADD domains. To establish a functional role of the ADD domain binding to unmodified H3 tails, we analyzed the DNA methylation of in vitro reconstituted chromatin with Dnmt3a2, the Dnmt3a2/Dnmt3L complex, and the catalytic domain of Dnmt3a. All Dnmt3a complexes preferentially methylated linker DNA regions. Chromatin substrates with unmodified H3 tail or with H3K9me3 modification were methylated more efficiently by full-length Dnmt3a and full-length Dnmt3a/3L complexes than chromatin trimethylated at H3K4. In contrast, the catalytic domain of Dnmt3a was not affected by the H3K4me3 modification. These results demonstrate that the binding of the ADD domain to H3 tails unmethylated at K4 leads to the preferential methylation of DNA bound to chromatin with this modification state. Our in vitro results recapitulate DNA methylation patterns observed in genome-wide DNA methylation studies.
Figures


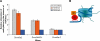
References
-
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. - PubMed
-
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. - PubMed
-
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

