A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair
- PMID: 20224553
- PMCID: PMC2868568
- DOI: 10.1038/emboj.2010.27
A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair
Abstract
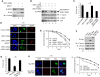
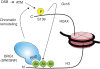
Although recent studies highlight the importance of histone modifications and ATP-dependent chromatin remodelling in DNA double-strand break (DSB) repair, how these mechanisms cooperate has remained largely unexplored. Here, we show that the SWI/SNF chromatin remodelling complex, earlier known to facilitate the phosphorylation of histone H2AX at Ser-139 (S139ph) after DNA damage, binds to gamma-H2AX (the phosphorylated form of H2AX)-containing nucleosomes in S139ph-dependent manner. Unexpectedly, BRG1, the catalytic subunit of SWI/SNF, binds to gamma-H2AX nucleosomes by interacting with acetylated H3, not with S139ph itself, through its bromodomain. Blocking the BRG1 interaction with gamma-H2AX nucleosomes either by deletion or overexpression of the BRG1 bromodomain leads to defect of S139ph and DSB repair. H3 acetylation is required for the binding of BRG1 to gamma-H2AX nucleosomes. S139ph stimulates the H3 acetylation on gamma-H2AX nucleosomes, and the histone acetyltransferase Gcn5 is responsible for this novel crosstalk. The H3 acetylation on gamma-H2AX nucleosomes is induced by DNA damage. These results collectively suggest that SWI/SNF, gamma-H2AX and H3 acetylation cooperatively act in a feedback activation loop to facilitate DSB repair.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
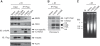





Similar articles
-
ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair.Oncogene. 2015 Jan 15;34(3):303-13. doi: 10.1038/onc.2013.556. Epub 2014 Jan 13. Oncogene. 2015. PMID: 24413084
-
Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction.EMBO J. 2006 Sep 6;25(17):3986-97. doi: 10.1038/sj.emboj.7601291. Epub 2006 Aug 24. EMBO J. 2006. PMID: 16932743 Free PMC article.
-
Genome-wide screen of human bromodomain-containing proteins identifies Cecr2 as a novel DNA damage response protein.Mol Cells. 2012 Jul;34(1):85-91. doi: 10.1007/s10059-012-0112-4. Epub 2012 Jun 12. Mol Cells. 2012. PMID: 22699752 Free PMC article.
-
Chromatin response to DNA double-strand break damage.Epigenomics. 2011 Jun;3(3):307-21. doi: 10.2217/epi.11.14. Epigenomics. 2011. PMID: 22122340 Review.
-
The role of the SWI/SNF chromatin remodelling complex in the response to DNA double strand breaks.DNA Repair (Amst). 2020 Sep;93:102919. doi: 10.1016/j.dnarep.2020.102919. DNA Repair (Amst). 2020. PMID: 33087260 Review.
Cited by
-
SWI/SNF recruitment to a DNA double-strand break by the NuA4 and Gcn5 histone acetyltransferases.DNA Repair (Amst). 2015 Jun;30:38-45. doi: 10.1016/j.dnarep.2015.03.006. Epub 2015 Mar 25. DNA Repair (Amst). 2015. PMID: 25869823 Free PMC article.
-
ARID1A regulates DNA repair through chromatin organization and its deficiency triggers DNA damage-mediated anti-tumor immune response.Nucleic Acids Res. 2024 Jun 10;52(10):5698-5719. doi: 10.1093/nar/gkae233. Nucleic Acids Res. 2024. PMID: 38587186 Free PMC article.
-
HSP110 Regulates the Assembly of the SWI/SNF Complex.Cells. 2025 Jun 5;14(11):849. doi: 10.3390/cells14110849. Cells. 2025. PMID: 40498025 Free PMC article.
-
β-catenin functions as a molecular adapter for disordered cBAF interactions.Mol Cell. 2025 Aug 21;85(16):3041-3056.e9. doi: 10.1016/j.molcel.2025.06.026. Epub 2025 Jul 21. Mol Cell. 2025. PMID: 40695292
-
The Transcriptional Coactivator ADA2b Recruits a Structural Maintenance Protein to Double-Strand Breaks during DNA Repair in Plants.Plant Physiol. 2018 Apr;176(4):2613-2622. doi: 10.1104/pp.18.00123. Epub 2018 Feb 20. Plant Physiol. 2018. PMID: 29463775 Free PMC article.
References
-
- Agalioti T, Chen G, Thanos D (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392 - PubMed
-
- Allard S, Masson JY, Cote J (2004) Chromatin remodeling and the maintenance of genome integrity. Biochim Biophys Acta 1677: 158–164 - PubMed
-
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y (2007) New nomenclature for chromatin-modifying enzymes. Cell 131: 633–636 - PubMed
-
- Bao Y, Shen X (2007) Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev 17: 126–131 - PubMed
-
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF (2002) Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

