The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex
- PMID: 20224578
- PMCID: PMC2854590
- DOI: 10.1038/embor.2010.12
The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex
Abstract
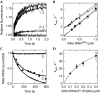
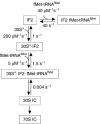
Bacterial translation initiation factor 2 (IF2) is a GTPase that promotes the binding of the initiator fMet-tRNA(fMet) to the 30S ribosomal subunit. It is often assumed that IF2 delivers fMet-tRNA(fMet) to the ribosome in a ternary complex, IF2.GTP.fMet-tRNA(fMet). By using rapid kinetic techniques, we show here that binding of IF2.GTP to the 30S ribosomal subunit precedes and is independent of fMet-tRNA(fMet) binding. The ternary complex formed in solution by IF2.GTP and fMet-tRNA is unstable and dissociates before IF2.GTP and, subsequently, fMet-tRNA(fMet) bind to the 30S subunit. Ribosome-bound IF2 might accelerate the recruitment of fMet-tRNA(fMet) to the 30S initiation complex by providing anchoring interactions or inducing a favourable ribosome conformation. The mechanism of action of IF2 seems to be different from that of tRNA carriers such as EF-Tu, SelB and eukaryotic initiation factor 2 (eIF2), instead resembling that of eIF5B, the eukaryotic subunit association factor.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M (2006a) How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol Cell 23: 183–193 - PubMed
-
- Brandi L, Fabbretti A, Milon P, Carotti M, Pon CL, Gualerzi CO (2007) Methods for identifying compounds that specifically target translation. Methods Enzymol 431: 229–267 - PubMed
-
- Canonaco MA, Calogero RA, Gualerzi CO (1986) Mechanism of translational initiation in prokaryotes. Evidence for a direct effect of IF2 on the activity of the 30S ribosomal subunit. FEBS Lett 207: 198–204 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

