Morphological and genomic characterization of Filobasidiella depauperata: a homothallic sibling species of the pathogenic cryptococcus species complex
- PMID: 20224779
- PMCID: PMC2835752
- DOI: 10.1371/journal.pone.0009620
Morphological and genomic characterization of Filobasidiella depauperata: a homothallic sibling species of the pathogenic cryptococcus species complex
Abstract
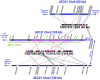
The fungal species Cryptococcus neoformans and Cryptococcus gattii cause respiratory and neurological disease in animals and humans following inhalation of basidiospores or desiccated yeast cells from the environment. Sexual reproduction in C. neoformans and C. gattii is controlled by a bipolar system in which a single mating type locus (MAT) specifies compatibility. These two species are dimorphic, growing as yeast in the asexual stage, and producing hyphae, basidia, and basidiospores during the sexual stage. In contrast, Filobasidiella depauperata, one of the closest related species, grows exclusively as hyphae and it is found in association with decaying insects. Examination of two available strains of F. depauperata showed that the life cycle of this fungal species shares features associated with the unisexual or same-sex mating cycle in C. neoformans. Therefore, F. depauperata may represent a homothallic and possibly an obligately sexual fungal species. RAPD genotyping of 39 randomly isolated progeny from isolate CBS7855 revealed a new genotype pattern in one of the isolated basidiospores progeny, therefore suggesting that the homothallic cycle in F. depauperata could lead to the emergence of new genotypes. Phylogenetic analyses of genes linked to MAT in C. neoformans indicated that two of these genes in F. depauperata, MYO2 and STE20, appear to form a monophyletic clade with the MATa alleles of C. neoformans and C. gattii, and thus these genes may have been recruited to the MAT locus before F. depauperata diverged. Furthermore, the ancestral MATa locus may have undergone accelerated evolution prior to the divergence of the pathogenic Cryptococcus species since several of the genes linked to the MATa locus appear to have a higher number of changes and substitutions than their MATalpha counterparts. Synteny analyses between C. neoformans and F. depauperata showed that genomic regions on other chromosomes displayed conserved gene order. In contrast, the genes linked to the MAT locus of C. neoformans showed a higher number of chromosomal translocations in the genome of F. depauperata. We therefore propose that chromosomal rearrangements appear to be a major force driving speciation and sexual divergence in these closely related pathogenic and saprobic species.
Conflict of interest statement
Figures








Similar articles
-
Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales.Eukaryot Cell. 2009 Mar;8(3):353-61. doi: 10.1128/EC.00373-08. Epub 2009 Jan 16. Eukaryot Cell. 2009. PMID: 19151324 Free PMC article.
-
The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insights into the evolution of sex and sex-determining chromosomal regions in fungi.PLoS Genet. 2010 May 20;6(5):e1000961. doi: 10.1371/journal.pgen.1000961. PLoS Genet. 2010. PMID: 20502678 Free PMC article.
-
Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex.PLoS Genet. 2012;8(2):e1002528. doi: 10.1371/journal.pgen.1002528. Epub 2012 Feb 16. PLoS Genet. 2012. PMID: 22359516 Free PMC article.
-
The Evolution of Sexual Reproduction and the Mating-Type Locus: Links to Pathogenesis of Cryptococcus Human Pathogenic Fungi.Annu Rev Genet. 2019 Dec 3;53:417-444. doi: 10.1146/annurev-genet-120116-024755. Epub 2019 Sep 19. Annu Rev Genet. 2019. PMID: 31537103 Free PMC article. Review.
-
Diversity of the Cryptococcus neoformans-Cryptococcus gattii species complex.Rev Iberoam Micol. 2008 Mar;25(1):S4-12. doi: 10.1016/s1130-1406(08)70019-6. Rev Iberoam Micol. 2008. PMID: 18338917 Review.
Cited by
-
Molecular and genetic evidence for a tetrapolar mating system in the basidiomycetous yeast Kwoniella mangrovensis and two novel sibling species.Eukaryot Cell. 2013 May;12(5):746-60. doi: 10.1128/EC.00065-13. Epub 2013 Mar 22. Eukaryot Cell. 2013. PMID: 23524993 Free PMC article.
-
Genetic and Genomic Analyses Reveal Boundaries between Species Closely Related to Cryptococcus Pathogens.mBio. 2019 Jun 11;10(3):e00764-19. doi: 10.1128/mBio.00764-19. mBio. 2019. PMID: 31186317 Free PMC article.
-
Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides.PLoS Pathog. 2011 Jun;7(6):e1002086. doi: 10.1371/journal.ppat.1002086. Epub 2011 Jun 16. PLoS Pathog. 2011. PMID: 21698218 Free PMC article.
-
Fungal evolution: major ecological adaptations and evolutionary transitions.Biol Rev Camb Philos Soc. 2019 Aug;94(4):1443-1476. doi: 10.1111/brv.12510. Epub 2019 Apr 25. Biol Rev Camb Philos Soc. 2019. PMID: 31021528 Free PMC article. Review.
-
Profiling a killer, the development of Cryptococcus neoformans.FEMS Microbiol Rev. 2012 Jan;36(1):78-94. doi: 10.1111/j.1574-6976.2011.00286.x. Epub 2011 Jul 4. FEMS Microbiol Rev. 2012. PMID: 21658085 Free PMC article. Review.
References
-
- Hurst LD, Pál CP, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nature Reviews. 2004;5:299–310. - PubMed
-
- Kahmann RR, Romeis T, Bolker M, Kamper J. Control of mating and development in Ustilago maydis. Curr Opin Genet Dev. 1995;5:559–564. - PubMed
-
- Kronstad JW, Staben C. Mating type in filamentous fungi. Annu Rev Genet. 1997;31:245–276. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

