The cold denatured state of the C-terminal domain of protein L9 is compact and contains both native and non-native structure
- PMID: 20225821
- PMCID: PMC3319020
- DOI: 10.1021/ja908104s
The cold denatured state of the C-terminal domain of protein L9 is compact and contains both native and non-native structure
Abstract
Cold denaturation is a general property of globular proteins, and the process provides insight into the origins of the cooperativity of protein folding and the nature of partially folded states. Unfortunately, studies of protein cold denaturation have been hindered by the fact that the cold denatured state is normally difficult to access experimentally. Special conditions such as addition of high concentrations of denaturant, encapsulation into reverse micelles, the formation of emulsified solutions, high pressure, or extremes of pH have been applied, but these can perturb the unfolded state of proteins. The cold denatured state of the C-terminal domain of the ribosomal protein L9 can be populated under native-like conditions by taking advantage of a destabilizing point mutation which leads to cold denaturation at temperatures above 0 degrees C. This state is in slow exchange with the native state on the NMR time scale. Virtually complete backbone (15)N, (13)C, and (1)H as well as side-chain (13)C(beta) and (1)H(beta) chemical shift assignments were obtained for the cold denatured state at pH 5.7, 12 degrees C. Chemical shift analysis, backbone N-H residual dipolar couplings, amide proton NOEs, and R(2) relaxation rates all indicate that the cold denatured state of CTL9 (the C-terminal domain of the ribosomal protein L9) not only contains significant native-like secondary structure but also non-native structure. The regions corresponding to the two native alpha-helices show a strong tendency to populate helical Phi and Psi angles. The segment which connects alpha-helix 2 and beta-strand 2 (residues 107-124) in the native state exhibits a significant preference to form non-native helical structure in the cold denatured state. The structure observed in the cold denatured state of the I98A mutant is similar to that observed in the pH 3.8 unfolded state of wild type CTL9 at 25 degrees C, suggesting that it is a robust feature of the denatured state ensemble of this protein. The implications for protein folding and for studies of cold denatured states are discussed.
Figures




 ) and I98A CTL9 (
) and I98A CTL9 ( ) at pH 5.7, 12 °C. A schematic diagram of the elements of secondary structure of the native state of CTL9 is shown at the top of each panel.
) at pH 5.7, 12 °C. A schematic diagram of the elements of secondary structure of the native state of CTL9 is shown at the top of each panel.
 ) for I98A CTL9 versus residue number. A schematic diagram of the elements of secondary structure of the native state of CTL9 is shown at the top of each panel.
) for I98A CTL9 versus residue number. A schematic diagram of the elements of secondary structure of the native state of CTL9 is shown at the top of each panel.
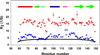
 ) and the cold denatured state of I98A CTL9 (
) and the cold denatured state of I98A CTL9 ( ). The black solid line (−) is the best fit to the phenomenological model of Schwalbe and coworkers. Data was collected at 12°C for both proteins.
). The black solid line (−) is the best fit to the phenomenological model of Schwalbe and coworkers. Data was collected at 12°C for both proteins.
Similar articles
-
The unfolded state of the C-terminal domain of the ribosomal protein L9 contains both native and non-native structure.Biochemistry. 2009 Jun 9;48(22):4707-19. doi: 10.1021/bi802299j. Biochemistry. 2009. PMID: 19301913 Free PMC article.
-
The low-pH unfolded state of the C-terminal domain of the ribosomal protein L9 contains significant secondary structure in the absence of denaturant but is no more compact than the low-pH urea unfolded state.Biochemistry. 2008 Sep 9;47(36):9565-73. doi: 10.1021/bi8006862. Epub 2008 Aug 16. Biochemistry. 2008. PMID: 18707127 Free PMC article.
-
Cooperative cold denaturation: the case of the C-terminal domain of ribosomal protein L9.Biochemistry. 2013 Apr 9;52(14):2402-9. doi: 10.1021/bi3016789. Epub 2013 Mar 26. Biochemistry. 2013. PMID: 23461364
-
The cold denatured state is compact but expands at low temperatures: hydrodynamic properties of the cold denatured state of the C-terminal domain of L9.J Mol Biol. 2007 Apr 20;368(1):256-62. doi: 10.1016/j.jmb.2007.02.011. Epub 2007 Feb 11. J Mol Biol. 2007. PMID: 17337003
-
Structural description of acid-denatured cytochrome c by hydrogen exchange and 2D NMR.Biochemistry. 1990 Nov 20;29(46):10433-7. doi: 10.1021/bi00498a001. Biochemistry. 1990. PMID: 2176867 Review.
Cited by
-
Temperature and urea have opposing impacts on polyproline II conformational bias.Biochemistry. 2013 Feb 5;52(5):949-58. doi: 10.1021/bi301435p. Epub 2013 Jan 27. Biochemistry. 2013. PMID: 23350874 Free PMC article.
-
Residual structure in unfolded proteins.Curr Opin Struct Biol. 2012 Feb;22(1):4-13. doi: 10.1016/j.sbi.2011.09.002. Epub 2011 Oct 4. Curr Opin Struct Biol. 2012. PMID: 21978577 Free PMC article. Review.
-
The Complex Energy Landscape of the Protein IscU.Biophys J. 2015 Sep 1;109(5):1019-25. doi: 10.1016/j.bpj.2015.07.045. Biophys J. 2015. PMID: 26331259 Free PMC article.
-
Effect of an Imposed Contact on Secondary Structure in the Denatured State of Yeast Iso-1-cytochrome c.Biochemistry. 2017 Dec 26;56(51):6662-6676. doi: 10.1021/acs.biochem.7b01002. Epub 2017 Dec 8. Biochemistry. 2017. PMID: 29148740 Free PMC article.
-
Observation of solvent penetration during cold denaturation of E. coli phosphofructokinase-2.Biophys J. 2013 May 21;104(10):2254-63. doi: 10.1016/j.bpj.2013.04.024. Biophys J. 2013. PMID: 23708365 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

