New statistical potential for quality assessment of protein models and a survey of energy functions
- PMID: 20226048
- PMCID: PMC2853469
- DOI: 10.1186/1471-2105-11-128
New statistical potential for quality assessment of protein models and a survey of energy functions
Abstract
Background: Scoring functions, such as molecular mechanic forcefields and statistical potentials are fundamentally important tools in protein structure modeling and quality assessment.
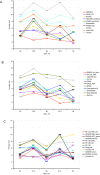
Results: The performances of a number of publicly available scoring functions are compared with a statistical rigor, with an emphasis on knowledge-based potentials. We explored the effect on accuracy of alternative choices for representing interaction center types and other features of scoring functions, such as using information on solvent accessibility, on torsion angles, accounting for secondary structure preferences and side chain orientation. Partially based on the observations made, we present a novel residue based statistical potential, which employs a shuffled reference state definition and takes into account the mutual orientation of residue side chains. Atom- and residue-level statistical potentials and Linux executables to calculate the energy of a given protein proposed in this work can be downloaded from http://www.fiserlab.org/potentials.
Conclusions: Among the most influential terms we observed a critical role of a proper reference state definition and the benefits of including information about the microenvironment of interaction centers. Molecular mechanical potentials were also tested and found to be over-sensitive to small local imperfections in a structure, requiring unfeasible long energy relaxation before energy scores started to correlate with model quality.
Figures


References
-
- Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal structures: quasi-chemical approximation. Macromolecules. 1985;18:534. doi: 10.1021/ma00145a039. - DOI
-
- Pohl FM. Empirical protein energy maps. NatNew Biol. 1971;234(52):277. - PubMed
-
- Samudrala R, Moult J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. JMolBiol. 1998;275(5):895. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

