Epigenetic histone modifications of human transposable elements: genome defense versus exaptation
- PMID: 20226072
- PMCID: PMC2836006
- DOI: 10.1186/1759-8753-1-2
Epigenetic histone modifications of human transposable elements: genome defense versus exaptation
Abstract
Background: Transposition is disruptive in nature and, thus, it is imperative for host genomes to evolve mechanisms that suppress the activity of transposable elements (TEs). At the same time, transposition also provides diverse sequences that can be exapted by host genomes as functional elements. These notions form the basis of two competing hypotheses pertaining to the role of epigenetic modifications of TEs in eukaryotic genomes: the genome defense hypothesis and the exaptation hypothesis. To date, all available evidence points to the genome defense hypothesis as the best explanation for the biological role of TE epigenetic modifications.
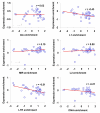
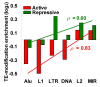
Results: We evaluated several predictions generated by the genome defense hypothesis versus the exaptation hypothesis using recently characterized epigenetic histone modification data for the human genome. To this end, we mapped chromatin immunoprecipitation sequence tags from 38 histone modifications, characterized in CD4+ T cells, to the human genome and calculated their enrichment and depletion in all families of human TEs. We found that several of these families are significantly enriched or depleted for various histone modifications, both active and repressive. The enrichment of human TE families with active histone modifications is consistent with the exaptation hypothesis and stands in contrast to previous analyses that have found mammalian TEs to be exclusively repressively modified. Comparisons between TE families revealed that older families carry more histone modifications than younger ones, another observation consistent with the exaptation hypothesis. However, data from within family analyses on the relative ages of epigenetically modified elements are consistent with both the genome defense and exaptation hypotheses. Finally, TEs located proximal to genes carry more histone modifications than the ones that are distal to genes, as may be expected if epigenetically modified TEs help to regulate the expression of nearby host genes.
Conclusions: With a few exceptions, most of our findings support the exaptation hypothesis for the role of TE epigenetic modifications when vetted against the genome defense hypothesis. The recruitment of epigenetic modifications may represent an additional mechanism by which TEs can contribute to the regulatory functions of their host genomes.
Figures



References
-
- Gould SJ, Vrba ES. Exaptation: a missing term in the science of form. Paleobiology. 1982;8:4–15.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

