Aptamer-targeted cell-specific RNA interference
- PMID: 20226078
- PMCID: PMC2835998
- DOI: 10.1186/1758-907X-1-4
Aptamer-targeted cell-specific RNA interference
Abstract
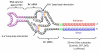
This potent ability of small interfering (si)RNAs to inhibit the expression of complementary RNA transcripts is being exploited as a new class of therapeutics for a variety of diseases. However, the efficient and safe delivery of siRNAs into specific cell populations is still the principal challenge in the clinical development of RNAi therapeutics. With the increasing enthusiasm for developing targeted delivery vehicles, nucleic acid-based aptamers targeting cell surface proteins are being explored as promising delivery vehicles to target a distinct disease or tissue in a cell-type-specific manner. The aptamer-based delivery of siRNAs can often enhance the therapeutic efficacy and reduce the unwanted off-target effects of siRNAs. In particular, for RNA interference-based therapeutics, aptamers represent an efficient agent for cell type-specific, systemic delivery of these oligonucleotides. In this review, we summarize recent attractive developments in creatively using cell-internalizing aptamers to deliver siRNAs to target cells. The optimization and improvement of aptamer-targeted siRNAs for clinical translation are further highlighted.
Figures

Similar articles
-
The therapeutic potential of cell-internalizing aptamers.Curr Top Med Chem. 2009;9(12):1144-57. doi: 10.2174/156802609789630893. Curr Top Med Chem. 2009. PMID: 19860714
-
Current progress on aptamer-targeted oligonucleotide therapeutics.Ther Deliv. 2013 Dec;4(12):1527-46. doi: 10.4155/tde.13.118. Ther Deliv. 2013. PMID: 24304250 Free PMC article. Review.
-
Targeted siRNA delivery using aptamer-siRNA chimeras and aptamer-conjugated nanoparticles.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019 May;11(3):e1543. doi: 10.1002/wnan.1543. Epub 2018 Aug 2. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019. PMID: 30070426 Review.
-
Development of cell-type specific anti-HIV gp120 aptamers for siRNA delivery.J Vis Exp. 2011 Jun 23;(52):2954. doi: 10.3791/2954. J Vis Exp. 2011. PMID: 21730942 Free PMC article.
-
Current progress of RNA aptamer-based therapeutics.Front Genet. 2012 Nov 2;3:234. doi: 10.3389/fgene.2012.00234. eCollection 2012. Front Genet. 2012. PMID: 23130020 Free PMC article.
Cited by
-
Aptamer-conjugated polymeric nanoparticles for targeted cancer therapy.Drug Deliv Transl Res. 2012 Dec;2(6):418-36. doi: 10.1007/s13346-012-0104-0. Drug Deliv Transl Res. 2012. PMID: 25787323
-
Chimeric aptamers in cancer cell-targeted drug delivery.Crit Rev Biochem Mol Biol. 2011 Dec;46(6):459-77. doi: 10.3109/10409238.2011.614592. Epub 2011 Sep 28. Crit Rev Biochem Mol Biol. 2011. PMID: 21955150 Free PMC article. Review.
-
Therapeutic potential of aptamer-siRNA conjugates for treatment of HIV-1.BioDrugs. 2012 Dec 1;26(6):393-400. doi: 10.2165/11635350-000000000-00000. BioDrugs. 2012. PMID: 23046156 Free PMC article. Review.
-
State-of-the-art gene-based therapies: the road ahead.Nat Rev Genet. 2011 May;12(5):316-28. doi: 10.1038/nrg2971. Epub 2011 Apr 6. Nat Rev Genet. 2011. PMID: 21468099 Review.
-
Nucleic acids in human glioma treatment: innovative approaches and recent results.J Signal Transduct. 2012;2012:735135. doi: 10.1155/2012/735135. Epub 2012 May 21. J Signal Transduct. 2012. PMID: 22685651 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
